1. Background
Bronchial asthma is the most common chronic airway disease and is the leading chronic childhood disease in most industrialized countries (1). According to the International Study of Asthma and Allergies in Children (ISAAC), the prevalence of asthma is 11.7% in 6 - 7 and 14.1% in 13 - 14 year age group (2). Increasing research suggests that asthma is a polygenetic hereditary disease. More than 100 asthma-related genes have been detected worldwide through positional cloning and genome-wide association studies. Orosomucoid 1-like protein 3 (ORMDL3) gene, a recently discovered asthma-related gene, is a member of the ORMDL gene family. It is localized on human chromosome 17q12-21. The ORMDL3 gene is highly conserved in yeasts and vertebrates, and encodes a transmembrane protein of 153 amino acids located in the endoplasmic reticulum (3). Maffatt et al first identified the ORMDL3 gene by genome-wide association study and demonstrated it was related to childhood-(younger than 16 years)-onset-asthma) (4). Other studies demonstrated that the ORMDL3 gene was also related to unfold protein responses (UPR) and intracellular sphingolipid metabolic balance (5-8). Its function extends to various inflammatory diseases and allergic diseases such as ulcerative colitis (9-12), ankylosing spondylitis and allergic asthma.
As molecular biology and molecular immunology techniques have progressed, cell signal transduction pathways have become hotspots for the study of the pathogenesis of asthma. STAT6 is a member of the signal transducer and activator of transcription (STATs) family. It is localized on human chromosome 17q11.1-22 and encodes a protein of 859 amino acids with five functional areas. Normally STAT6 is located in the cytoplasm and is expressed in all tissues in an inactive latent monomer. STAT6 undergoes phosphorylation to form dimers and then traffics into the nucleus to combine with DNA, and is inactivated by re-phosphorylation (13). The Janus kinase (JAK)/STAT6 signaling pathway plays a role in the pathogenesis of asthma. The upstream stimulating factors of JAK/STAT6 signaling pathway such as interleukin (IL)-4 and IL-13 are important cytokines involved in the pathogenesis of asthma. IL-4 and IL-13 are regulated by various receptor systems and take part in the activation of the enzyme Janus to cause STAT6 phosphorylation and its trafficking into the nucleus to regulate the transcription of its target genes (14). Our unpublished study demonstrated that transfection of STAT siRNA into human bronchial epithelial cells decreased the activity of the ORMDL3 promoter. Therefore, we hypothesized that IL4 and IL-13 are associated with STAT6 during the regulation of ORMDL3.
Glucocorticoids are the most effective drugs to control airway inflammation. Clinical and laboratory data reveal that intravenous administration or inhalation of glucocorticoids could influence multiple steps of asthma directly or indirectly, thus reducing the airway hyperresponsiveness, inflammation, and rate of asthma incidence, and improving lung function to decrease asthma-related mortality. Dexamethasone, a long-lasting glucocorticoid, inhibited the formation of IgE+ cells, mast cells and eosinophils induced by allergens (15). Dexamethasone also inhibited the secretion of multiple cytokines such as IL-4 and IL-13, thus inhibiting inflammation. Therefore, we speculated that dexamethasone might regulate the expression of ORMDL3 via the JAK/STAT6 signaling pathway.
2. Objectives
We investigated the expression of ORMDL3 and STAT6 in murine models of asthma. The correlations between IL-4, IL-13, STAT6, and ORMDL3 in mice were further confirmed. In addition, the effect of dexamethasone on the expression of IL-4, IL-13, STAT6, and ORMDL3 was also studied to provide a new theoretical basis for the treatment of asthma.
3. Methods
3.1. Ethical Statement
The study protocols were conducted with the approval of the institutional committee of Nanjing medical university. All animal experiments were performed in accordance with the guide for the care and Use of laboratory animals (national academy press, 1996).
3.2. Animals
Six-week-old female BALB/c mice were obtained from the experimental animal center of Nanjing medical university (Jiangsu, China) and were used in this study. A total of 30 mice were separated into three groups: control group (n = 10), untreated asthma group (asthma group, n = 10) and dexamethasone-treated group (Dex group, n = 10).
3.3. Asthma Protocols
The asthma group and Dex group were sensitized with 200 μL sensitization liquid [2 μg ovalbumin (OVA, A5503, Sigma-Aldrich, St. Louis, MO) mixed with 2 mg aluminum hydroxide (239186, Sigma-Aldrich, St. Louis, MO)] on days 1, 7 and 14 followed by challenge with 1% OVA for 30 min in a chamber on days 21 - 27. The Dex group was treated with dexamethasone (1 mg/kg, D4902, Sigma-Aldrich, St. Louis, MO) via intraperitoneal injection 30 minutes before sensitization. The control group was sensitized and challenged with normal saline instead of OVA. Twenty-four hours after the last challenge, the peripheral blood of mice from each group was obtained via the retro-orbital plexus. The blood samples were then centrifuged (1000 × g, 5 min, 4°C) and the plasma was stored at -80°C for the detection of IL-4 and IL-13. Mice were sacrificed by cervical dislocation under complete anesthesia and lungs were removed. The left lung was infused with 4% paraformaldehyde for histology and the right lung was stored in liquid nitrogen for real-time PCR and western blot analysis.
3.4. Histological and Immunohistochemistry Assessment
The left lungs of each group were embedded in paraffin and stained with hematoxylin and eosin (H and E). For immunohistochemical analysis, paraffin-embedded sections were dewaxed and cut into 4 μm slides. The slides were then incubated with primary antibodies against p-STAT6 (ab125308, Abcam, Cambridge, UK) and ORMDL3 (SC161143, Santa Cruz biotechnology, Santa Cruz, CA) overnight at 4°C. Then avidin/biotin/horseradish peroxidase complex (K0355, Dako Cytomation, Glostrup, Denmark) was added to the slides followed by 3, 3-diaminobenzidine (DAB) staining and hematoxylin counterstaining.
3.5. Enzyme-Linked Immunosorbent Assay (ELISA) Detection of Serum IL-4 and IL-13
Serum levels of IL-4 and IL-13 were detected by ELISA according to the manufacturer’s instructions (ab100710/ ab100700 Abcam, Cambridge, UK). The lower detection limits of these assays were 0.819 pg/mL (IL-4) and 8.3 pg/mL (IL-13). All measurements were performed in duplicate.
3.6. RNA Extraction and Quantitative Real-Time PCR
Total RNAs of tissue specimens from right lungs were extracted using Trizol reagent (Invitrogen, Carlsbad, CA ), and RNA was converted into cDNA using the PrimeScript RT Master Mix Perfect Real Time kit (RR036A, Takara, Otsu, Japan). Quantitative real-time PCR was performed on the Step One Plus Quantitative Real-Time PCR System (Applied Bio-Systems) using SYBR Green (Takara, Otsu, Japan). The mRNA expressions of mouse STAT6, ORMDL3, and β-actin were measured. The specific primer pairs are as follows:
STAT6: sense: 5’-CCCTGTGGGTTTGAACTCCTG-3’;
antisense: 5’-GTCGAAGTGAACCCGCTTCTG-3’;
ORMDL3: sense: 5’-GGGGGTGGTCAGGAAAGAGGCT-3’;
antisense: 5’-GGGTTGCCAGGAAGCCCACAAA-3’;
β-actin: sense: 5’-AACAGTCCGCCTAGAAGCAC-3’;
antisense: 5’-CGTTGACATCCGTAAAGACC-3’.
3.7. Western Blotting
Proteins were extracted from tissue specimens using the total protein extraction kit (KGP250, Keygentec, Nanjing, China) and protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA). Proteins were transferred onto SDS-PAGE gels and the blots were incubated with primary antibodies against p-STAT6 (ab125308, Abcam, Cambridge, UK), STAT6 (ab44718, Abcam, Cambridge, UK) and β-actin (Santa Cruz Biotechnology). Chemiluminescence signals were visualized and quantified using an ECL imager and Quantity One software (Bio-Rad).
3.8. Statistical Analysis
All data are expressed in mean ± standard deviation. Statistical analysis was conducted using SPSS version 22.0 (SPSS, Chicago, IL, USA). Group comparisons were accomplished with one-way ANOVA and P values < 0.05 were considered statistically significant.
4. Results
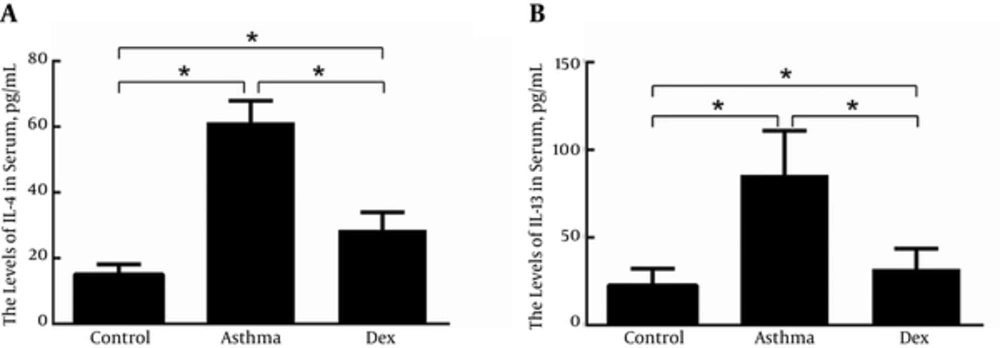
4.1. Determination of Serum IL-4 and IL-13
Serum IL-4 and IL-13 levels were measured by ELISA to validate the murine asthma model we established was successful and evaluate the efficacy of dexamethasone (Figure 1A and B). The concentrations of serum IL-4 and IL-13 in the asthma group were significantly higher than in the control group. In the Dex group, serum IL-4 and IL-13 concentrations were lower than in the asthma group but slightly higher than in the control group.
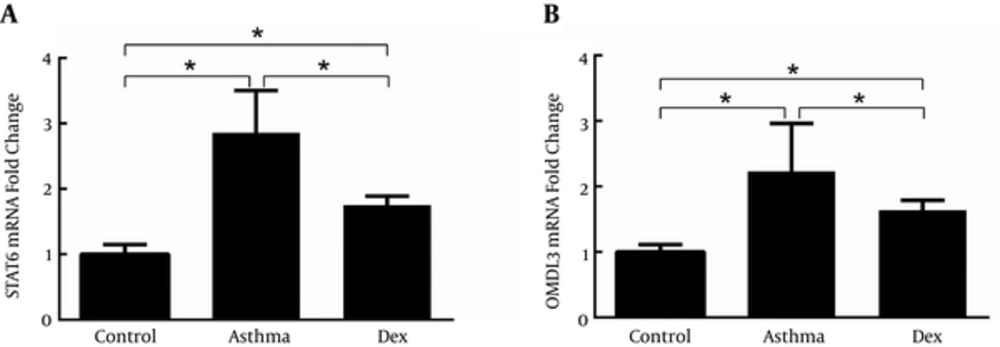
4.2. STAT6 and ORMDL3 Expression in Lungs
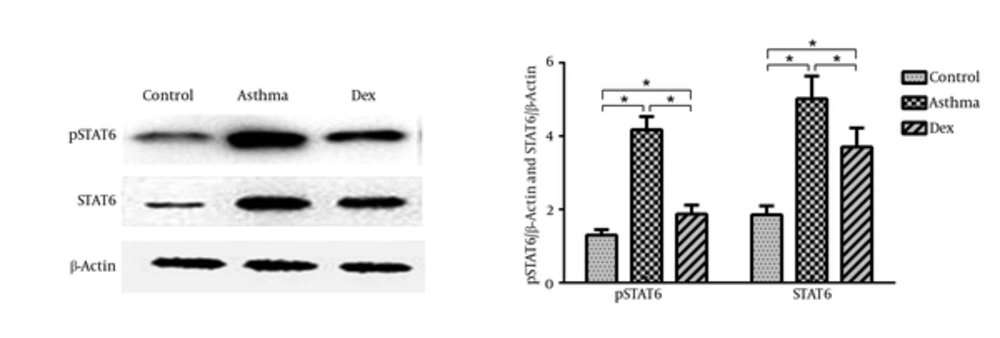
STAT6 and ORMDL3 mRNA expression was determined by quantitative real-time PCR (Figure 2A and 2B). Compared with the control group, the mRNA expression of both STAT6 and ORMDL3 was markedly increased by disease induction and both were suppressed by treatment with dexamethasone. The expression of STAT6 and p-STAT6 was measured by western blotting and showed an increase of STAT6 protein levels in the asthma group compared with the control group, which was suppressed by treatment with dexamethasone (Figure 3).
4.3. Histology and Immunohistochemistry
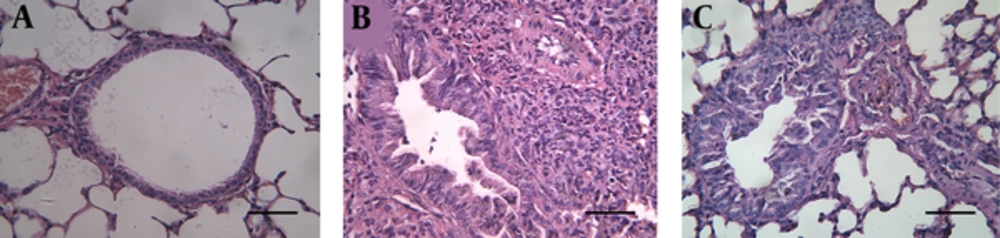
H&E staining revealed the structures of bronchi and alveoli were clear and integral without epithelial necrosis and inflammatory cellular infiltration in the control group (Figure 4A). In contrast, the asthma group contained high levels of eosinophil, lymphocyte, and plasma cell infiltration around the terminal bronchi and blood vessels (Figure 4B). Bronchial wall thickening and occlusions, epithelial necrosis and bronchospasm were also observed in the asthma group. All these pathological changes were partially suppressed by treatment with dexamethasone in the Dex group (Figure 4C).
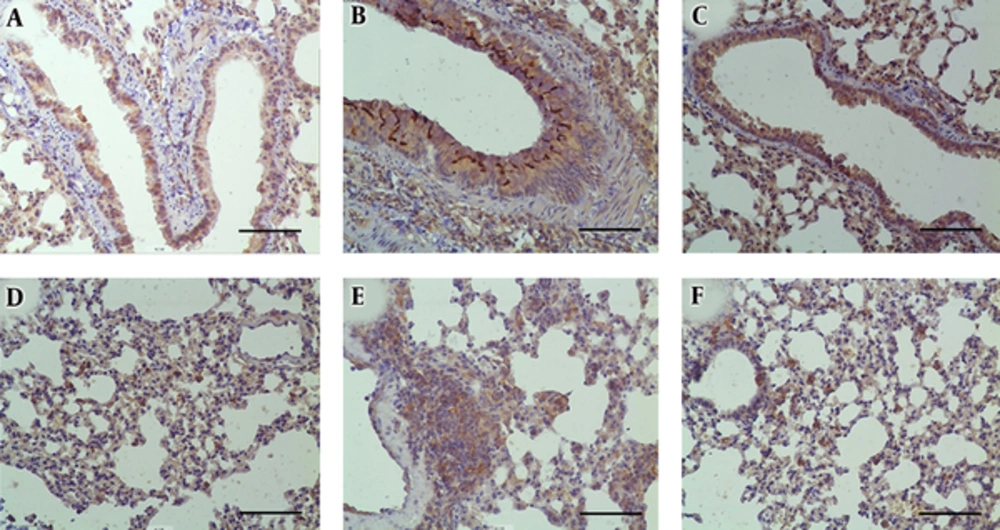
p-STAT6 staining showed expression mainly in airway epithelial cells and some inflammatory cells around the airway in all three groups (Figure 5A-C). The intensity of p-STAT6 staining was more significant in the asthma group (Figure 5B). Treatment with dexamethasone reduced p-STAT6 expression (Figure 5C). ORMDL3 staining demonstrated it was expressed in alveolar epithelial cells and significantly increased by disease induction (Figure 5E) and suppressed by dexamethasone treatment (Figure 5F).
Expression of p-STAT6 and ORMDL3 in lung tissues was detected by staining with anti-p-STAT6 and anti-ORMDL3 antibodies. The expression of p-STAT6 in the (a) control, (b) asthma, and (c) Dex groups. The expression of ORMDL3 in the (d) control, (e) asthma, and (f) Dex groups. Scale bars: 200 μm.
5. Discussion
ORMDL3 is a newly discovered asthma-related gene and its exact regulatory mechanisms remain unknown. Studies indicated that ORMDL3 might inhibit the endoplasmic reticulum Ca2+ pump and facilitate UPR, the endogenous inducer of inflammatory responses related to the antibody secretion of plasma cells, lymphocyte development, and activity of c-Jun N-terminal kinase. This may be an important mechanism of ORMDL3 in the induction of asthma (16). Another mechanism might be related to sphingolipids because Orm proteins are critical mediators of sphingolipid homeostasis, which is related to immune cell migration, mast cell degranulation and airway hyperresponsiveness (7). A correlation study of 1411 Asian children revealed that GSDMB/ORMDL3 variants might induce bronchial hyperresponsiveness and asthma, where eosinophils mediated this process (17). The current study is the first to reveal that GSDMB/ORMDL3 polymorphisms might play an important role in the progress of eosinophil-mediated bronchial hyperresponsiveness which is considered to be the key points in the pathophysiology of asthma. We also observed a marked increase of ORMDL3 mRNA expression and high eosinophil infiltration of lung tissues from the asthma group compared with the control group consistent with previous research. To show the level of ORMDL3 proteins, immunohistochemistry was performed and showed a similar result as for the other techniques used.
Recently, there has been an increase in research on the regulation mechanisms of the ORMDL3 gene. Qiu et al indicated that the -64 to -56 bp region of the ORMDL3 gene promoter contained a STAT6 binding site which could combine with STAT6 directly and regulate the expression of human ORMDL3 (18). In our study, STAT6 mRNA and protein expression was also tested. We showed that there was a significant increase in STAT6 mRNA and protein expression in the asthma group compared with the control group. In addition, p-STAT6 protein expression was analyzed by immunohistochemistry, demonstrating that disease induction promoted p-STAT6 protein expression in mouse lung. We suspected that STAT6 signal pathways were activated in asthmatic mice. Miller et al indicated that allergen challenge induced ORMDL3 expression in wild-type mouse epithelium in vitro and in vivo and that the administration of IL-4 or IL-13 intranasally induced the significant expression of ORMDL3 mRNA in bronchial epithelium (6). Our result also showed that the levels of serum IL-4 and IL-13 were much higher in the asthma group compared with controls. A previous study demonstrated that the JAK/STAT6 pathway was activated by IL-4/IL-13, which might induce Th2 predominant immune responses in asthma (19). STAT6 may up-regulate immuno-related genes by combination with the promoter of target genes, thus regulating immune responses. When JAK/STAT6 signaling pathways are activated the expression levels of IL-4, IL-13 and IL-5 in mouse bronchoalveolar lavage fluid were significantly increased and Th2 responses were predominant. The degree of Th2 activity was positively correlated with the severity of asthma. Th2 predominant immune responses were inhibited in STAT6-/- mice and stimulation with IL-4 or IL-13 did not induce B-cells to produce IgE (20-23). Based on these results, our research in an asthma animal model further confirmed that the expression of ORMDL3 was possibly regulated by IL4, IL-13, and STAT6. Dexamethasone is a first-line drug for controlling bronchial asthma attack, and decreases airway hyperresponsiveness and inflammatory reactions. Our research showed that dexamethasone intervention could alleviate airway hyperresponsiveness, and reduce the serum levels of IL-4 and IL-13 and the expression of STAT6 and ORMDL3 in lung tissues. Therefore, dexamethasone might inhibit the activation of the JAK/STAT6 signaling pathway by reducing the serum levels of IL-4 and IL-13, thus inhibiting STAT6 phosphorylation and binding to the promoter region of ORMDL3. This process ultimately resulted in downregulating the expression of ORMDL3.
5.1. Conclusions
This study demonstrated that the serum levels of IL-4 and IL-13 and the expression of STAT6 and ORMDL3 in lung tissues were markedly increased in murine models of asthma, and significantly suppressed by dexamethasone treatment. Thus, the newly discovered asthma-related gene, ORMDL3, may become a new target for the diagnosis and treatment of asthma. Further studies are needed to identify which signal pathways are associated with the ORMDL3 regulation of immune responses and how ORMDL3 interacts with environmental and individual factors.