1. Background
Many studies have shown a significant relationship between intestinal bacteria and autism spectrum disorder (ASD) (1, 2). The microbiome-gut-brain axis is suggested to have a vital role in the development of ASD (3-5). Dysbiotic gut microbiota of children with ASD may be associated with brain dysfunction, but the underlying mechanisms are unclear. Furthermore, no specific gut bacteria are considered a biomarker for ASD so far. However, the symptom of some ASD cases has been improved statistically by therapies targeting intestinal bacteria (3, 4). Differences in biomarkers from various research studies might be linked to demographic characteristics of subjects such as region and race. Alcaligenaceae, Enterobacteriaceae, and Clostridium were considered candidate bacterial biomarkers by Li et al. (6). Another study showed gut bacterial biomarkers, such as Eubacterium, Bifidobacterium, Blautia, and Dialister (7). There are few reports about the intestinal microbiome of ASD in China. Furthermore, ASD initiates in the early life of children (8), while very little literature focuses on the early life of autistic children in China.
Vitamin A (VitA) is an important micronutrient for people. Researchers have recently reported that VitA plays a role in intestinal bacteriomes (9). Amit-Romach et al. suggested that VitA also regulates intestinal bacillus (10). Another study revealed that neonatal vitamin A supplementation and vitamin A status were associated with the composition of the intestinal microbiome among infants at 2 years old (11). Some studies have shown the relationship between vitamin A and ASD (12-14). Studies also showed that individuals with ASD have lower VitA concentrations than typically developing (TD) children. Recent studies have reported that serum VA level was negatively correlated with the severity of ASD (15). However, little is reported on how vitamin A regulates specific gut microbiota to affect ASD.
2. Objectives
This study further explored the characteristics of intestinal bacteria in ASD in China. In addition, the effect of VitA on the gut bacteria of ASD was investigated to find a new possible treatment for ASD.
3. Methods
Ten children with ASD aged 2 to 6 from special training facilities in Chongqing from 2017 to 2018 were enrolled in this study. According to the 5th edition of the diagnostic and statistical manual of mental disorders (DSM-5), the inclusion criteria included children diagnosed by more than two child behavioral health practitioners. The exclusion criteria included children diagnosed with other congenital disorders. Those with acute and chronic infectious diseases in the past month were also excluded. The other exclusion criteria were dietary supplements taken by children during the past six months and probiotics and antibiotics taken for almost a month. At the same time, 10 healthy children aged 2 to 6 years were enrolled as controls from a kindergarten between 2017 and 2018. The exclusion criteria were dietary supplements taken by children during the past six months and probiotics and antibiotics taken for almost a month.
A demographic questionnaire was used to obtain information, including gender, age, nationality, and living place. The study was consented to by parents (guardians) of all children, and we obtained the approval of the Committee of Chongqing Medical and Pharmaceutical College.
The dietary VitA intake was investigated using three-day 24-hour dietary recalls (validity, 0.73; reliability, 0.69) (16, 17), and the data were analyzed by a nutrition Software program (FeiHua V2.5, Beijing, China). Serum retinal analysis was detected by high-performance liquid chromatography (HPLC) (18).
The fresh and pollution-free stool was collected and temporarily stored in a refrigerator at -20°C and was transferred to the refrigerator at -80°C within 7 days.
The 16S rRNA genes of distinct regions (16SV4) were amplified using a specific primer (16S V4: 515F - 806R) with the barcode. Sequencing was performed by Uparse software (Uparse v70.1001) (19). Qiime software (version 1.9.1) was used to calculate the observed species, Chao1, Shannon, Simpson, and abundance-based coverage estimator (ACE) (20). The linear discriminant (LDA) analysis effect size (LEfSe) software was used for LEfSe analysis, and the filter value of the LDA score was set at the default value of 4.
Data were analyzed by SPSS 17.0 software package. The p values below 0.05 were significant.
4. Results
4.1. Demographic Characteristics
All children with ASD were 2 to 6 years old in the ASD group (8 males and 2 females; 57.50 ± 11.38 months on average). The control group contained 10 healthy children aged 2 to 6 (8 males and 2 females; 50.50 ± 11.22 months on average). There was no statistical difference between the two groups concerning gender and age (P > 0.05). All the children were of Han nationality and lived in Chongqing City.
4.2. Vitamin A in Diet and Serum
Vitamin A dietary intakes were 185.00 ± 38.12 and 261.70 ± 79.70 μgRE in the ASD and control groups, respectively. There were 5 cases (50%) in the ASD group that had an intake of lower than dietary nutrient reference intake (DRIS), according to the Chinese Academy of Nutrition, while there were two such cases (30%) in the control group. In addition, the retinol level in serum was 0.78 ± 0.24 μmol/L in the ASD group and 1.01 ± 0.24 μmol/L in the control group. VitA deficiency (VAD) was observed in 3 cases (70%) in the ASD group and 5 cases (50%) in the control group. The two groups significantly differed in dietary VitA intake and serum VitA levels (P < 0.05).
4.3. Characteristics of Gut Bacteria
4.3.1. Alpha Diversity Analysis
We obtained an average of 82,261 sequences and 619 operational taxonomic units (OUTs) per sample (Figure 1). The alpha diversity analysis revealed no significant differences using the Simpson. However, the scores of observed species (reflecting species richness), Chao, and ACE (reflecting species abundance) (Table 1) regarding the OTU levels in autistic samples decreased compared to those in controls, suggesting that autistic children had less species diversity. The findings revealed that the overall composition of the ASD gut microbiota differed from that of the control gut microbiota.
| Control Group | ASD Group | P Value | |
|---|---|---|---|
| Observed species | 595 | 515 | 0.0008 |
| Shannon | 5.534 | 4.913 | 0.0292 |
| Simpson | 0.918 | 0.895 | 0.3363 |
| Chao1 | 641.163 | 565.026 | 0.0011 |
| ACE | 655.215 | 582.327 | 0.0017 |
Alpha Diversity Analysis in Two Groups
4.3.2. Beta Diversity Analysis
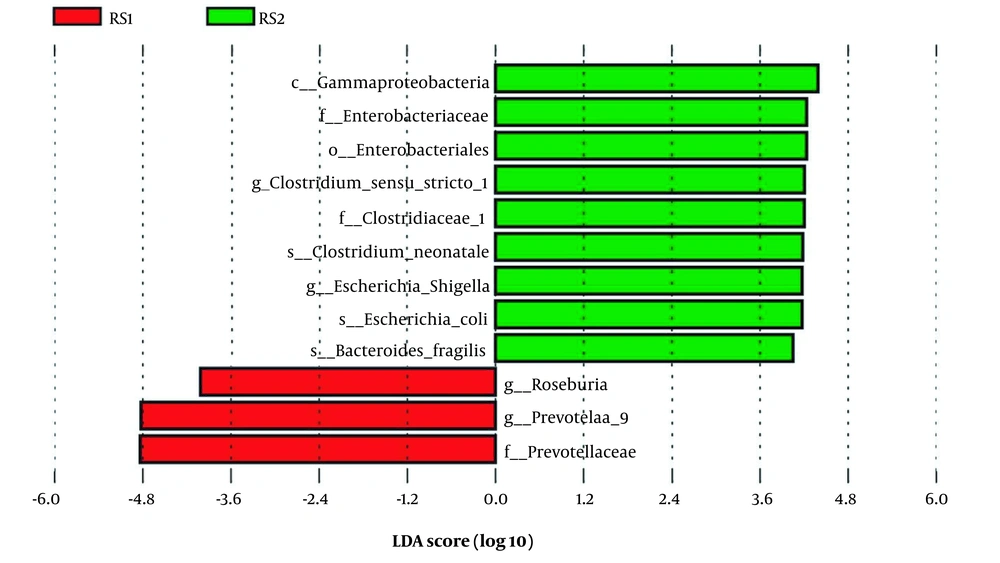
The LEFSe analysis can identify biomarkers with statistical differences between the groups. This study found increasing relative abundance in Prevotellaceae (family level), Prevotella 9, and Roseburia at the genus level in the ASD group. However, significantly lower abundances were observed in 9 bacteria (Enterobacteriales (order level), Gammaproteobacteria (class level), Enterobacteriaceae (family level), Clostridiaceae 1 (family level), Clostridium sensu stricto 1 (genus level), Escherichia-Shigella (genus level), Bacteroides fragilis (species level), Escherichia coli (species level), and Clostridium neonatale (species level) (Figure 2).
4.3.3. Vitamin A is Associated with Intestinal Bacteria
The analysis found that the level of serum retinol was associated negatively with the abundance of Prevotella 9 (r = -0.708, P = 0.022), while there was no relationship between retinol level and the abundance of other bacteria which were considered biomarkers in this study (Table 2).
| R | P | |
|---|---|---|
| f_Prevotellaceae | -0.48 | 0.165 |
| g_Prevotella 9 | -0.71 | 0.022 |
| g_Roseburia | 0.35 | 0.327 |
Correlation Between Serum Retinol and Gut Bacteria in Children with Autism Spectrum Disorder
5. Discussion
In the present study, less species diversity and altered relative abundance of gut bacteria were observed in children with ASD from Chongqing, China, consistent with previous reports (21) to some extent.
There was an obvious increase in Prevotella at the family level, Prevotella 9, and Roseburia at the genus level in autistic children in our research. Several studies have demonstrated significant enrichment of Prevotella and Roseburia in the intestinal microbiome of children with ASD (22, 23). Studies also suggested the potential role of Prevotella as intestinal pathobionts (24). A report showed that some Prevotella strains may participate in human disease by promoting chronic inflammation (25). However, some reported that children with ASD showed a depletion of Prevotella, and microbial metastasis therapy improved autism-related symptoms and the abundance of Prevotella (26, 27). The different properties of Prevotella in the gut microbiome between these studies might be due to the high genetic diversity within and between the species (24). Further study is needed to explore the differences between their results and ours.
Moreover, autistic children displayed a reduction in nine bacteria, which is somewhat consistent with some studies, but different from others (23, 28, 29). Decreased Bacteroides fragilis was found in our study, consistent with previous reports (30, 31). Meanwhile, there was a lower abundance of Escherichia coli and Escherichia-Shigella in the intestinal microbiome of children with ASD in this study, similar to the literature (32, 33). The Clostridium sensu stricto 1 (genus level) and Clostridium neonatale (species level) were found to be lower in the intestinal bacteria of children with ASD in this study, while Clostridia spp. increased in ASD children (2). Further studies are needed to investigate the role of Clostridia in ASD. All studies strongly indicate that the microbiome-gut-brain axis is important in health and disease. Intestinal bacteria have been found to play a major role in the development of autism, although there have been no specific biomarkers yet, and the real causes leading to the development of autism are still unclear.
Furthermore, this study found a negative correlation between serum VitA and intestinal Prevotella in children with ASD, suggesting that vitamin A may regulate intestinal bacteria. Huda et al. reported that early infant supplementation of vitamin A could increase the abundance of Bifidobacterium in the intestines of boys (11). However, few reports have shown the relationship between VitA and Prevotella gut bacteria in autistic children. Our study proposes that VitA may affect the development of ASD by regulating the Prevotella of gut bacteria.
5.1. Limitations
The sample size was insufficient, which may lead to difficulty distinguishing true differences from noise in this study. Nevertheless, based on the existing methodology, we will be able to enlarge the sample for further study in the future. Meanwhile, this study was limited to children aged 3 to 6. Whether the results apply to autistic children of other age groups will be confirmed by expanding age groups in the future. In fact, this experiment selecting 3 to 6 years old children aimed to eliminate age interference on the results and make the results more reliable.
5.2. Conclusions
Our study helps identify some bacterial biomarkers for ASD, as in previous reports. Differing from some other research, increased Prevotella abundance was found in this study, which might be due to some reasons, such as the high genetic diversity within and between the species. Further study about the different results between those and ours is needed.
Meanwhile, the study suggests that dietary VitA may be involved in the clinical symptoms of ASD by regulating the intestinal bacteria Prevotella. It may provide a new way to treat ASD in the future. Further studies will be needed to identify the results by expanding the sample size and developing animal experiments.