1. Background
Heroin addiction presents a significant public health challenge. Methadone maintenance treatment (MMT) is the primary treatment option. However, MMT's effectiveness varies due to genetic and environmental factors. Some patients experience treatment failure, relapse, or developing tolerance (1). The complexity of heroin addiction's neurobiology involves various neurotransmitter systems, notably the dopaminergic and opioidergic systems (2).
Dopamine receptors (DRs) (DRD1, DRD2, DRD3, DRD4, and DRD5) and the COMT enzyme, which degrades dopamine, play crucial roles in addiction. Opioid receptors (ORs) (OPRM1, OPRD1, and OPRK1) are directly impacted by heroin, affecting mood regulation and stress responses (3).
Brain-derived neurotrophic factor (BDNF), vital for neuronal growth and synaptic plasticity, experiences changes in heroin users, influencing reward pathways and cravings (4). Additionally, RAB22A and DNM1L are involved in cellular processes, such as trafficking and mitochondrial fission, respectively, which can indirectly relate to the neural adaptations observed in addiction (5).
Methadone, by action on mu-opioid receptors, modulates dopamine release more consistently than heroin, thereby impacting DRs and contributing to long-term adaptive changes in the brain. These changes can influence MMT outcomes, including mood disturbances and the severity of withdrawal symptoms, as quantified by tools, such as the Subjective Opiate Withdrawal Scale (SOWS) (6).
The primary issue that remains unresolved in the treatment of heroin addiction through MMT is its varied effectiveness, largely influenced by genetic and environmental factors (1). Given the complexities of heroin addiction, combined with the existing knowledge gap, there is a clear need for an initial screening panel before initiating MMT.
2. Objectives
The present study delved into how a combination of specific genes and demographic variables could potentially influence and predict an individual's level of compliance with MMT for heroin addiction. The ultimate goal is to develop a model that can forecast a patient's likelihood of successfully adhering to MMT. To explore these questions, an adoption design study was proposed by identifying potential biomarkers using non-invasive techniques, specifically through peripheral blood sampling. Initially, the focus was on identifying any changes in the gene expression profiles related to addiction pathogenesis pathways during MMT. Subsequently, the study examined a correlation between genetic profiles and environmental risk factors, in addition to their interactions in response to methadone. It is hypothesized that by identifying key genetic and personal factors, a pre-treatment screening tool can be created. This tool would enable a more tailored approach to MMT, aligning treatment strategies more closely with each patient's unique genetic profile and personal setting, thereby enhancing the overall effectiveness of heroin addiction management.
3. Methods
3.1. Sample Study
This cross-sectional study was conducted on 80 heroin users and 80 age- and gender-matched healthy controls between 2019 and 2020 to assess the study objectives. The group of genes that display the most powerful evidence for positive correlation with addiction pathogenesis was selected based on preceding findings by others and previous work of our group (5, 7, 8).
The first group of patients comprised compliance to treatment with a methadone daily dose of 40 mg/d or greater to 80 mg/d or less (good compliance [GC] to MMT, n = 58). The second group of patients comprised poor compliance (PC) to treatment with a methadone daily dose of 120 mg/d or greater and who were receiving no co-medications known to be inducers of methadone metabolism (PC to MMT, n = 22). Most of the sociodemographic and clinical data were collected from the clinical records (Appendix 1 in the Supplementary File provides sample classification methodology and inclusion criteria, Appendix 2 in the Supplementary File, demographic table).
3.2. Gene Expression
Primers were designed using Primer Express software (http://www.ncbi.nlm.ih.gov/tools/primer-blast/) (Appendix 3 in the Supplementary File). In this study, 5 mL of blood was used for peripheral blood mononuclear cells (PBMC) separation using Ficoll-Hypaque (Pharmacia, Sweden). The lymphocytes' messenger ribonucleic acid (mRNA) was extracted using a Roche RNA mini kit (Germany), and complementary deoxyribonucleic acid (cDNA) was synthesized by Fermentas cDNA synthesis kit (Germany). Real-time polymerase chain reaction (PCR) was performed using a Techne Flexigene PCR Cycler (USA) and LightCycler® FastStart DNA MasterPLUS SYBR Green I kit (Roche, Germany).
3.3. Statistical Analysis
Real-time PCR data were analyzed using LinRegPCR (version 2021.2) and normalized against the β-actin gene. Unpaired t-test (P < 0.05) and graphical representations were performed in GraphPad Prism 10.
Data analysis was conducted in SPSS-27 using the Kolmogorov-Smirnov test, Wilcoxon’s test, Levene's test, and effect size (Cohen's d, Hedges' g, Glass’s Δ). Linear, hierarchical, and logistic regressions were employed for predictive modeling. Multivariate logistic regression with backward elimination identified variables associated with MMT compliance, used for propensity matching. The predictive power of biomarkers was assessed via receiver operating characteristic (ROC) curves and area under the curve (AUC) evaluation (P < 0.05), and the Hosmer-Lemeshow test verified model accuracy.
4. Results
4.1. Gene Expression Associated with Heroin Addiction
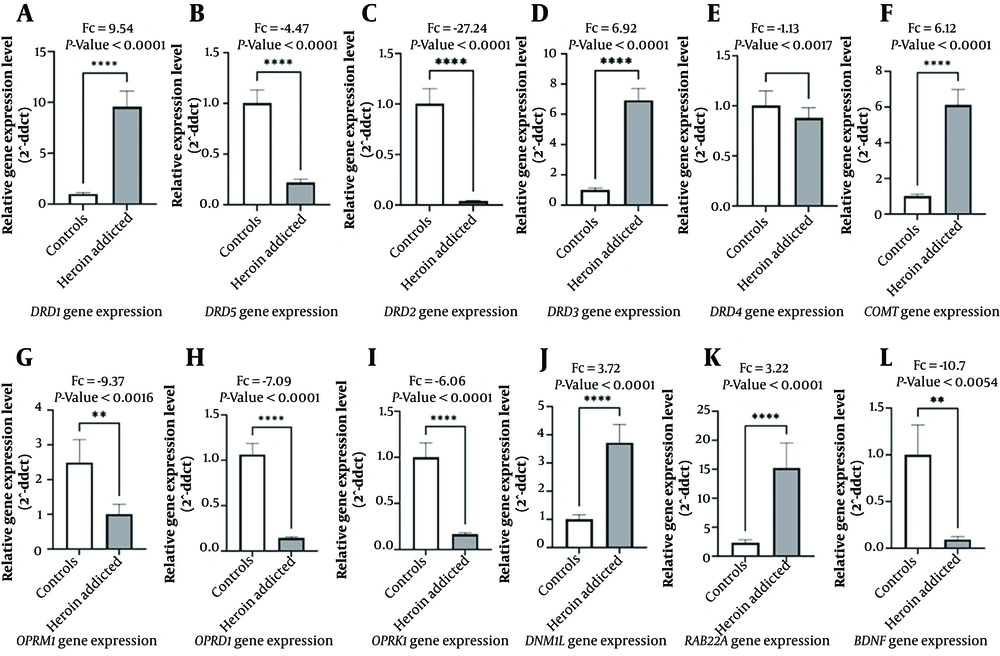
The Mann-Whitney U test between heroin-addicted and controls revealed a marked decrease in OPRM1 in the addicted group (P < 0.0016, Fc: -9.37). Nevertheless, DNM1L and RAB22A showed increased expression, with DNM1L (P < 0.0001, Fc 3.72) and RAB22A (P < 0.0001, Fc 3.22).
The unpaired t-test in DRs (DRD1-DRD5) revealed that DRD1 increased 9.54-fold; however, DRD2 decreased by -27.24-fold in the addicted group (P < 0.0001). The COMT gene expression increased 6.12-fold, and ORs (OPRD1 and OPRK1) decreased (Fc -7.09 and -6.06, respectively, P < 0.0001). Additionally, BDNF gene expression significantly decreased in the addicted group (Fc -10.7, P < 0.0054) (Appendix 4 in the Supplementary File) (Figure 1).
4.2. Gene Expression in MMT
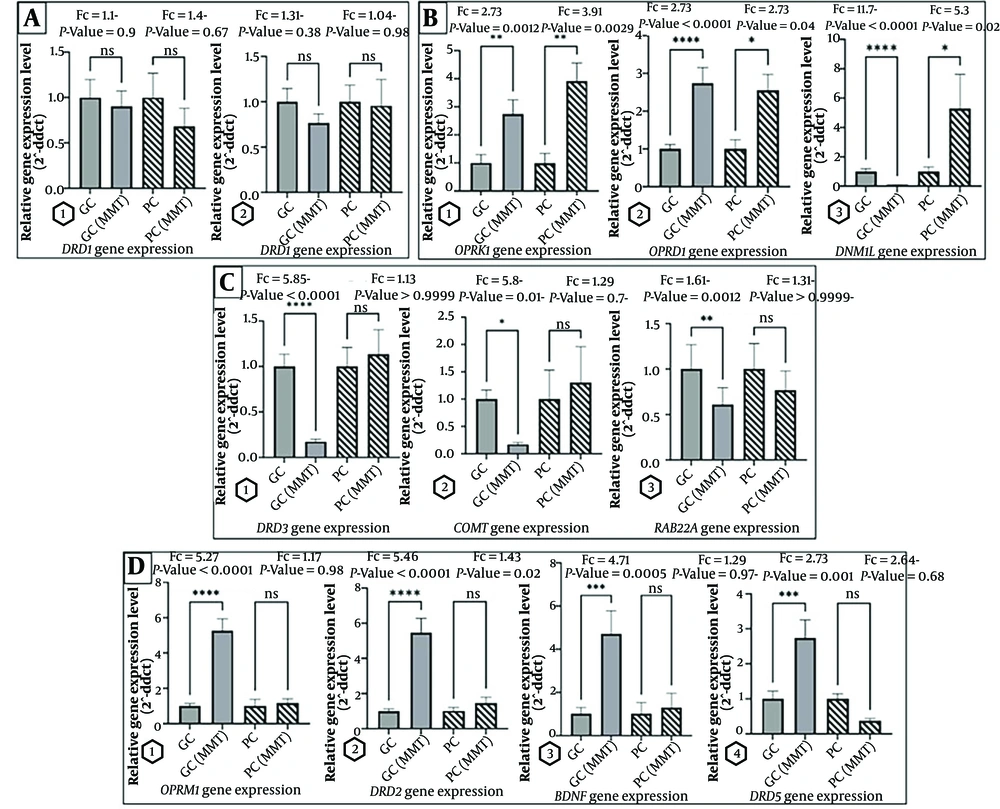
In the GC-MMT, DRD3, COMT, and RAB22A gene expression decreased (Fc -5.85, -5.8, and -1.61 respectively). In contrast, the OPRM1, OPRD1, OPRK1, DRD2, BDNF, and DRD5 genes increased, (Fc 5.27, 2.73, 2.73, 5.46, 4.71, and 2.73 respectively). The aforementioned findings underscore the varied impact of methadone treatment compliance on different gene expressions in addiction (Appendix 5 in the Supplementary File, Figure 2).
4.3. Correlation of Gene Expression with Methadone Dose
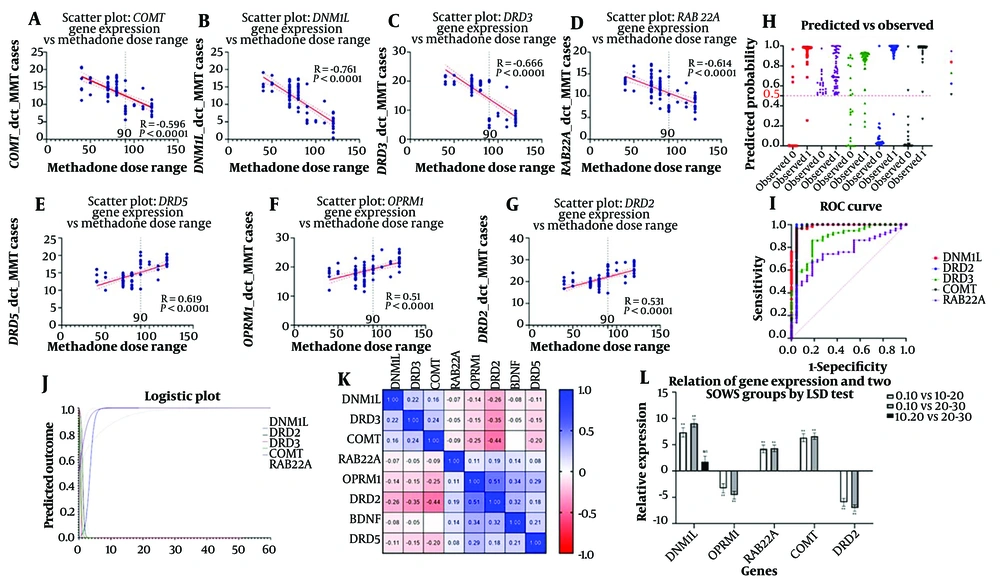
OPRM1 exhibited a significant positive correlation (r = 0.540, P < 0.0001), and RAB22A showed a significant negative correlation (r = -0.614, P < 0.0001) with methadone dose, particularly at higher doses.
Moreover, Spearman correlation analysis showed strong negative correlations for COMT (r = -0.596, P < 0.0001) and DNM1L (r = -0.761, P < 0.0001) with methadone dose, especially significant in higher dose ranges. DRD2 and DRD5 demonstrated significant positive correlations (DRD2: r = 0.531, P < 0.0001; DRD5: r = 0.619, P < 0.0001), mainly in higher doses. DRD3 revealed a strong negative correlation in higher doses (r = -0.666, P < 0.0001) (Appendix 6 in the Supplementary File) (Figure 3A-G).
(A-G) Scatter plots illustrate the significant correlations between the normalized gene expression levels (ΔCT) and the range of methadone doses administered during MMT. (H) The scatter plot depicts the predicted probabilities of patient compliance to methadone treatment against the actual observed outcomes. Predicted probabilities were derived from a logistic regression model, with individual observations represented by symbols. (I) The ROC curve graph evaluates the diagnostic ability of five genetic markers to predict methadone treatment compliance. (J) This logistic plot depicts the relationship between the probability of the predicted outcome and the values of independent genetic variables. (K) The matrix displays the pairwise Pearson correlation coefficients (R2 values) among eight genes studied in the context of methadone treatment response. (L) This bar graph depicts the relative gene expression levels associated with two categories of the subjective SOWS scores, comparing low (0 - 10) to high (20 - 30) withdrawal symptom scores. Statistical significance determined by the least significant difference (LSD) test is denoted above the bars: ** P < 0.01, * P < 0.05, ns indicates not significant.
4.4. Predictive Validity of Genetic Markers in Compliance to Methadone
For each of the DNM1L, DRD3, COMT, RAB22A, OPRM1, DRD2, BDNF, and DRD5 gene expressions, a separate simple binary logistic regression was conducted where the predictor was the gene expression, and the outcome was compliance to methadone. In a binary logistic regression analysis, the increased DNM1L expression was significantly associated with PC (93.75% model accuracy); however, higher DRD2 expression correlated with GC (97.50% model accuracy). The COMT gene expression showed a substantial influence on compliance, with 96.25% predictive accuracy and a significant odds ratio (Appendix 7 in the Supplementary File). The scatter plot (Figure 3H), ROC curve (Figure 3I), logistic plot (Figure 3J), and correlation matrix (Figure 3K) highlighted the significant results.
Moderate correlations were observed between OPRM1 and DRD2, suggesting potential multicollinearity issues in regression models. To mitigate this, genes were carefully selected based on their biological relevance and clinical significance.
Differential gene expression profile between GC and PC implies a genetic predisposition influencing MMT efficacy, especially in higher dose ranges. This study also extended to demographic factors using analysis of variance (ANOVA) tests, emphasizing the importance of considering both genetic and demographic influences on MMT's effectiveness.
4.5. Gene Variations and Demographic Influences on MMT
The study evaluated compliance levels with MMT based on clinical guidelines for withdrawal management and considered various independent variables, such as age, addiction history, heroin use duration, and SOWS scores.
The analysis of variance analysis revealed significant gene expression differences related to SOWS scores, particularly in DNM1L and OPRM1, indicating varying gene expression with withdrawal severity (Appendix 8 in the Supplementary File, Figure 3L). Effect sizes highlighted strong associations for DNM1L and DRD3 (Appendix 9 in the Supplementary File).
Further ANOVA tests examined gene expression against heroin use duration, showing significant impacts on genes, such as DRD2. This analysis revealed varying effect sizes, suggesting differential gene expression impacts based on heroin dose (Appendix 10 in the Supplementary File).
This study also investigated the relationship between heroin use onset and gene expression (Appendix 11 in the Supplementary File). Significant differences were observed for DNM1L, OPRM1, and DRD2, indicating a strong link between the age of onset and gene expression.
Lastly, this study employed hierarchical logistic regression, focusing on genetic and demographic predictors for non-compliance with MMT, setting the stage for more detailed multiple logistic regression analyses. The binary logistic regression model's validity was confirmed by Omnibus Tests of Model Coefficients (P < 0.0001), and Hosmer-Lemeshow test (chi-square = 0.000, P = 1.000).
Multiple logistic regression demonstrated excellent performance, with an area under the ROC curve of 0.9945 (Table 1). DNM1L, COMT, DRD2, negative family history, and lower heroin doses showed positive coefficients, indicating their influence on increasing compliance likelihood.
| Parameter | Estimate | Standard Error | 95% CI | Odds Ratio | 95% CI (Odds Ratio) | Z-Value | P-Value | VIF | R² with Other Variables |
|---|---|---|---|---|---|---|---|---|---|
| Intercept (β0) | -44.16 | 17.1 | -90.91 to -17.68 | 6.61E-20 | 3.29E-040 to 2.09E-08 | 2.582 | 0.0098 | - | - |
| DNM1L a (β1) | 1.134 | 0.4383 | 0.4430 to 2.292 | 3.108 | 1.557 to 9.899 | 2.587 | 0.0097 | 9.138 | 0.8906 |
| COMT a (β2) | 1.238 | 0.5059 | 0.4982 to 2.693 | 3.449 | 1.646 to 14.78 | 2.448 | 0.0144 | 6.862 | 0.8543 |
| DRD2 a (β3) | 0.4542 | 0.2308 | 0.04824 to 1.051 | 1.575 | 1.049 to 2.861 | 1.968 | 0.049 | 10.53 | 0.905 |
| Family history b [0] (β4) | 4.076 | 1.998 | 0.6705 to 9.184 | 58.9 | 1.955 to 9742 | 2.04 | 0.0413 | 1.864 | 0.4635 |
| Heroin dose c [1] (β5) | 2.471 | 1.257 | 0.4136 to 6.229 | 11.83 | 1.512 to 507.4 | 1.966 | 0.0493 | 6.523 | 0.8467 |
a Inputted data for gene value was delta CT
b Family history [0] = Negative; [1]: Positive
c Heroin dose categories: [1]: 0.5 - 1.4/ [2]: 1.5 - 2.4/ [3]: Above 2.5
Model diagnostics affirmed the robustness of the analysis, with high accuracy in classification (97.5%). The predictive formula derived from the model is as follows:
ρ = (e-44.16 + 1.134 × DNM1L + 1.238 × COMT + 0.4542 × DRD2 + 4.076 × Patients Family History [0] + 2.47 × Heroin Dose [1]) / (1 + e-44.16 + 1.134 × DNM1L + 1.238 × COMT + 0.4542 × DRD2 + 4.076 × Patients Family History [0] + 2.47 × Heroin Dose [1])
This equation estimates the probability of compliance to MMT, integrating both genetic predispositions and drug use behavior, refined for the prediction in a clinical setting.
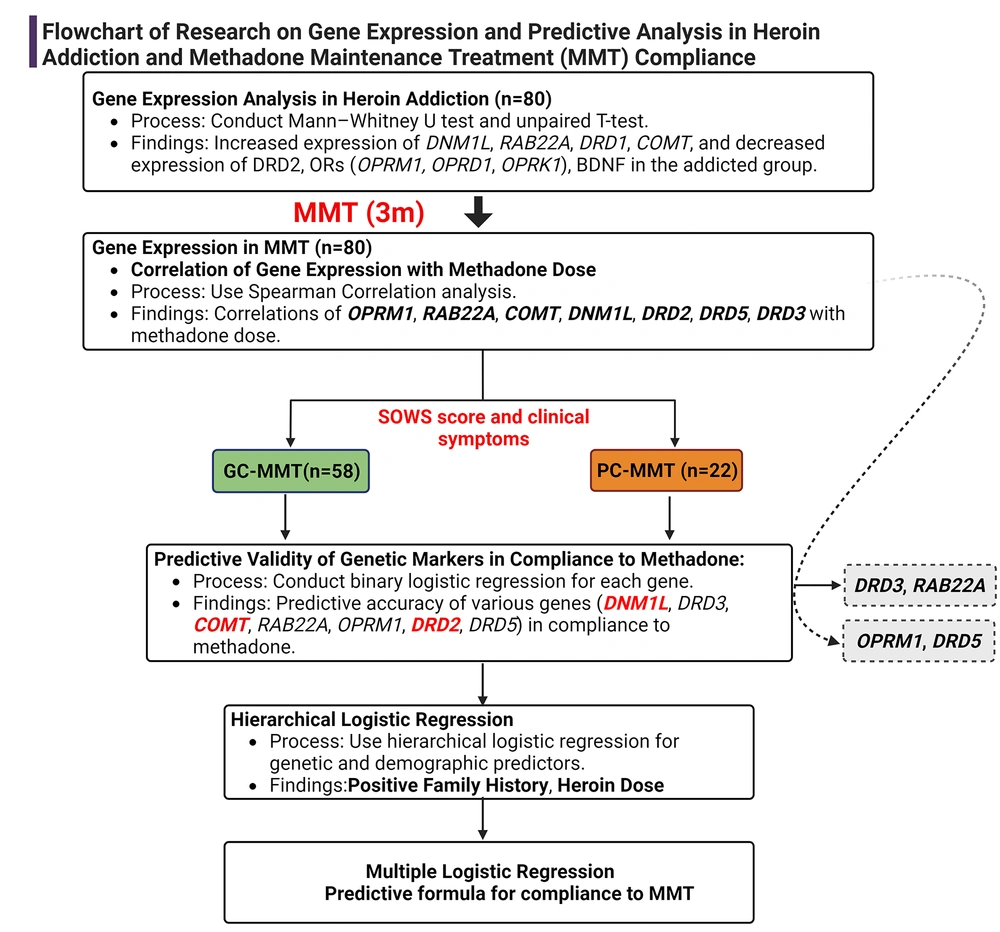
In summary, the present study's workflow visually summarizes the research (Figure 4), presenting a clear and concise representation of the complex interplay between gene expression and compliance with MMT.
Workflow summarizing gene expression's role in MMT compliance, from analysis to predictive modeling. Created with BioRender.com.
5. Discussion
Heroin addiction is a complex condition influenced by the combination of genetic elements and environmental factors (9). This study highlighted distinct variations in gene expression when analyzing the gene profile associated with heroin. Opioid receptor genes, along with DRD2, DRD5, and BDNF, showed decreased expression in heroin addiction; however, DRD1, DRD3, COMT, DNM1L, and RAB22A were upregulated. The decreased OR expression suggests reduced sensitivity, impacting pain management and addiction treatment (10). The COMT gene showed varied expression among heroin addicts. The DRD1 and DRD3 gene variations were linked to early-onset heroin dependence, indicating susceptibility to addiction (11). Previous research indicated conflicting findings regarding DRD5 and DRD4 (12). The majority of the studies focused on the brain’s expression. Therefore, quantifying DRD4 and DRD5 in peripheral blood is challenging due to low abundance, highlighting the complexities of studying these genes outside the central nervous system (CNS). The genetic susceptibility to heroin addiction was strongly associated with DRD2, evident in high expression levels in blood and particular polymorphisms (13). Studies also highlighted the influence of cocaine and morphine on DNM1L and RAB22A (5, 14); nevertheless, research on their expression in heroin addicts is missing. Additionally, alterations in BDNF expression in blood were proposed as a possible biomarker for heroin addiction treatment.
Regarding MMT, GC was associated with increased expression of ORs, BDNF, DRD2, and DRD5. Nonetheless, PC showed no significant change, except for OPRD1 and OPRK1. Research has yielded inconsistent interpretations regarding the relationship between OPRM1 expression and methadone tolerance (15), suggesting the influence of OPRM1 expression on methadone tolerance is multifaceted and might not be a direct factor.
Addiction has been shown to impact the epigenetic mechanism, such as promoter hypermethylation, leading to altered expression of ORs (16). Methadone maintenance treatment showed positive outcomes by increasing the expression of OPRM1 (17). Nevertheless, there was resistance to normalizing OPRM1 in PC-MMT.
To seek an explanation, this study examined the relationship between methadone dosage and patient categorization. Lower methadone doses consistently administered in GC led to increased OPRM1 expression, and different dosage groups (40 - 59.9 mg, 60 - 89.9 mg, and 90 mg or higher) showed varied gene expression and response patterns, indicating potential threshold effect at 90 mg where therapeutic responses change significantly, highlighting the complex nature of dose-dependent efficacy and tolerance.
The findings of the present study suggest a significant link between the age of initial heroin use, the presence of a positive family history, and OPRM1 gene expression, with early adolescence being a critical period for heroin initiation that might affect tolerance development in MMT. This finding is consistent with the higher heroin dosages observed in PC that necessitate increased methadone dosages in this subgroup. Moreover, this finding aligns with previous reports indicating that starting heroin use at an older age correlates with more effective outcomes from MMT (18), highlighting the importance of genetic predispositions in MMT outcomes.
Previous studies indicated that stressors can change BDNF expression in addiction-related brain areas (19), potentially inducing depressive symptoms in prolonged heroin use (20). Elevated BDNF levels have been observed to be linked to addictive behaviors (21). This study showed no notable change in BDNF expression in PC-MMT, contradicting prior beliefs about its link to MMT. This finding aligns with recent studies (22); however, the inconsistent results suggest that BDNF might not be a reliable biomarker for MMT efficacy.
The DRD2 gene expression remains unchanged in PC-MMT. Furthermore, lower DRD2 expression correlates with nonresponsiveness to MMT and higher prior heroin use. Methadone has been shown to increase DRD2 expression in the blood, potentially impacting the brain's reward system (23). The present study showed that higher methadone doses are associated with lower DRD2 gene expression in the PC group. These insights collectively suggest that DRD2 expression might be a pivotal factor in determining treatment resistance in opioid addiction, necessitating consideration of both genetic and environmental factors in tailoring MMT (24). Several pharmacogenetic studies have further emphasized the relationship between genetic factors and MMT response (23), highlighting DRD2's role in individual variances in pharmacotherapy response and methadone dosage requirements (25).
In addition, a significant decrease was noted in the expression of DNM1L, DRD3, COMT, and RAB22A genes in GC-MMT. On the other hand, those with PC-MMT did not exhibit substantial changes in gene expression, except for DNM1L, which intriguingly showed an increase. Additionally, changes in the DRD1 and DRD4 genes were not significant in both the GC and PC groups.
Currently, this is the first study regarding the direct connection between the RAB22A and DNM1L genes expression in peripheral blood and their implications for heroin addiction and the efficacy of methadone treatment. Notably, most research concerning RAB22A gene expression has been focused on its association with various types of cancers (26). The RAB22A gene, which is a part of the RAS oncogene family and plays a significant role in intracellular trafficking, might play a role in receptor endocytosis, contributing to resistance in addiction.
The logistic regression analysis revealed the complex interplay between genetic, environmental, and behavioral influences on MMT compliance. A significant finding is a positive coefficient for DNM1L, with an odds ratio of 3.108, underscoring its substantial role in MMT compliance. This gene, known for its involvement in mitochondrial dynamics and cellular energy regulation, has previously been linked to the therapeutic response in depression, where its upregulation was observed (27). Neurological evidence further supports a bidirectional relationship between heroin addiction and depression (28). Multiple studies suggest a connection between inflammation, mitochondrial dysfunction, increased reactive oxygen species (ROS) production, and the pathophysiology of depression (29). Additionally, the presence of depression in heroin addicts has been shown to influence methadone dosing, often requiring higher doses for stabilization in those with comorbid mood disorders (28). This is further supported by the observation that heroin-dependent individuals, once tolerant, are driven by a need to maintain homeostasis, which is linked to reduced dopamine release in key brain areas and an inability to inhibit drug-seeking behaviors (30).
As previously mentioned, another predictor was DRD2 based on the previous evidence (31) and observations of DRD2 downregulation's association with reduced MMT efficacy.
The COMT's role in breaking down dopamine has been linked to psychiatric disorders often co-existing with addiction. Higher COMT enzyme activity and reduced frontal cortical dopamine are more prevalent in heroin-dependent individuals (31). The present study further observed a connection between lower COMT expression and better MMT compliance.
The logistic model demonstrated significant associations of COMT and DRD2 with MMT response, with odds ratios of 3.449 and 1.575, respectively. In the concept of environmental and behavioral factors, the analysis reveals a pronounced effect of positive family history and higher heroin dose on response to MMT. Particularly, the high odds ratio for negative family history (58.9) highlights its substantial protective impact, potentially overshadowing other factors. Similarly, the heroin dose presents a considerable odds ratio of 11.83 for the lowest level as a protective factor and emphasizes its critical role in compliance with treatment.
The findings of the current study suggest that genetic factors significantly influence compliance with MMT, while environmental and behavioral factors, such as family history and heroin dosage, also play key roles. Individuals with a family history of addiction often show more severe opioid dependence symptoms (9). Family-related risk factors, including prenatal exposure to maternal smoking and inadequate care, lower parental education, neglect, and exposure to substance use within the family, contribute to the risk of drug abuse (32). It is believed that in the family risk factor, both the family environment and genetic predispositions might increase susceptibility to drug abuse.
Research indicates that higher methadone doses can reduce heroin use and alleviate withdrawal symptoms, suggesting their effectiveness in creating cross-tolerance to heroin (33). However, this relationship is complex (34), as higher doses might not always suppress withdrawal symptoms, particularly in patients with poor compliance to treatment. The effectiveness of methadone dosage varies based on individual responses and the importance of consistent treatment strategies.
This predictive model can guide clinicians in tailoring MMT strategies. Specifically, patients with no family history of addiction, lower heroin abuse, a lower expression level of COMT and DNM1L genes, and an increased expression level of the DRD2 gene typically show better results and adherence to MMT and might benefit from a modified approach. Nevertheless, deviations from this pattern tend to result in poorer compliance with MMT, which could lead to more side effects and less effective treatment.
Based on the current logistic regression model, this study has developed a predictive framework that can estimate an individual's likelihood of adhering to MMT, considering their genetic makeup, family history, and initial heroin dose. This area of study is still evolving, and further research is needed to fully understand these mechanisms. These insights could be instrumental in developing targeted strategies for management, prevention, or treatment in pre-selecting the most appropriate curation strategy.
4.1. Limitations and Future Directions
The current study on MMT compliance offers insights but has some limitations. The three-month follow-up might not capture long-term outcomes, and logistic regression analysis might oversimplify complex MMT compliance relationships. While significant P-values show strong associations, causality is not established, and findings might not be widely generalizable due to the specific population and moderate sample size. Additionally, gene expression measurements were performed in blood samples, not directly reflecting brain processes (35). Furthermore, more diverse research is needed for a deeper understanding and personalized treatment strategies.
4.2. Conclusions
This study showed that a positive family history, higher heroin usage, increased expression levels of COMT and DNM1L, and decreased expression of DRD2 are associated with poor compliance with MMT outcomes. These findings represent a significant step in the application of personalized medicine strategies in addiction treatment, offering a novel approach to optimizing MMT by considering individual genetic and environmental profiles.