1. Background
Drug abuse is an important national and social health problem and a high-risk factor for other diseases (1). It has been estimated that nearly 1.12 million Iranian people have drug abuse disorder (2). Opioid dependence is a chronic relapsing disorder (3). There are approximately 16 million illicit opioid-dependent individuals worldwide (4). The opioid-dependent individuals deal with many different health problems including infectious diseases such as HIV and hepatitis infection (5). Therefore, opioid substitution treatment as a harm reduction program is offered as an important strategy to decrease the consequences of drug abuse, provide significant social benefits, and improve public health.
Methadone is widely administered for opioid substitution treatment (6). The long-standing maintenance treatment by methadone was endorsed by the World Health Organization (WHO), UNODC, and UNAIDS (Joint United Nations Program on HIV/AIDS) in 2004 that argues a significant relationship between substitution therapy and substantial reductions in drug abuse, criminal activities, overdose, and conditions leading to a high risk of HIV infection (7). Methadone treatment success highly depends upon the optimal prescribed dosage (8). Therefore, to achieve better long-term outcomes, personalized therapy is required.
A few factors such as age, sex, and pathology have shown a link to the methadone dosage requirement. Importantly, genetic variability is now identified as a vital factor in determining optimal drug use and efficacy (9). Broad pharmacogenomics research has shown genetic variations in drug receptors, transporters, and metabolizing enzymes such as cytochrome P450s (CYPs) (10). P-glycoprotein is the product of the ABCB1 (MDR1) gene that belongs to the membrane transporters superfamily of adenosine triphosphate-binding cassette (11). It is an efflux drug transporter with a wide range of substrates that is widely expressed in different tissues including the central nervous system (CNS) and contributes to drug absorption, distribution, and elimination (12). Moreover, P-glycoprotein expression in the CNS modulates the opioids penetration and access to their action in the blood-brain barrier (13). The P-glycoprotein expression level and functional integrity influence its kinetic interaction with administered drugs and hence, play an important role in drug treatment success (14).
ABCB1 is a highly polymorphic gene bearing at least 50 single nucleotide polymorphisms (SNPs) located in both exonic and intronic regions of the gene. The most common polymorphisms identified in white populations are found in exon 2 (A61G), exon 11 (G1199A), exon 12 (C1236T), exon 21 (G2677T), and exon 26 (C3435T) (15). Although evidence shows the effect of many SNPs in the ABCB1 gene on methadone response, because of a high interpersonal variation of ABCB1 sequence (16), the haplotypes surrounding ABCB1 SNPs may be better predictors of P-glycoprotein function. Despite many reports on the frequency of ABCB1 SNPs in white populations, to our knowledge, there is no report of the prevalence of ABCB1 genetic variability in Iranian populations.
2. Objectives
Therefore, in the present study, we aimed to identify the frequency of ABCB1 SNPs and haplotypes in Iranian opioid-dependent individuals and the relationship between ABCB1 haplotypes and P-glycoprotein function.
3. Materials and Methods
3.1. Subjects
This cross-sectional study was conducted on 400 patients undergoing methadone treatment. They were randomly selected from methadone maintenance treatment (MMT) clinics in Shahroud, Semnan province, Iran. We used a multi-stage method for randomized sampling in which, the patients were selected by simple random sampling from some of the clinics as random clusters using a basket of numbered balls. From the selected clinics, 320 people who were under treatment for at least six months and had received a constant dose in the last two months with no underlying illness or other substance abuses were sorted according to their doses. Individuals with a dose of greater than 100 mg/day and less than 60 mg/day were selected and divided into two groups. Finally, 169 opioid-dependent individuals were enrolled and classified into low-dose dependent (low-dose dependent (LDD), n = 86) and high-dose dependent (high-dose dependent (HDD), n = 83) categories based on the daily methadone use of < 60 mg/day for the LDD group and > 100 mg/day for the HDD group.
To calculate the sample size for comparing the averages or ratios between several groups, we considered the mean values obtained in previous studies, the power of 90%, and the confidence limit of 95% to estimate the sample size of 40 people for each group. As the sample size is estimated based on a quantitative variable that turns into a qualitative variable and in order to take into account the dropout rate, we approximately doubled the estimated sample size. The demographic data such as sex, age, and ethnicity were obtained for all participants as presented in Table 1. The subjects with at least six months of MMT were recruited from the MMT clinics in Shahroud, Semnan province, Iran. All MMT subjects had reached a stable maintenance treatment phase, the dosage of which had been separately optimized and was stable for more than two months. We excluded individuals receiving drug treatment including digoxin, fexofenadine, indinavir, vincristine, colchicine, topotecan, rifampin, ritonavir, cyclosporine, verapamil, erythromycin, ketoconazole, itraconazole, quinidine, or clarithromycin. Moreover, the subjects with any other chronic diseases were excluded from the study. Liver function tests were performed for all the enrolled individuals. The abnormal serum levels of SGOT, SGPT, and alkaline phosphatase were considered as exclusion criteria. Participants provided written informed consent before blood sample collection. All stages, including designing, sampling, laboratory tests, and data analysis, were conducted between May and October 2018. This study was approved by the Ethics Committee of Shahroud University of Medical Sciences under the code IR.SHMU.REC.1394.24.
| Characteristics | HDD Group | LDD Group | P Value |
|---|---|---|---|
| Liver function test (SGOT) | 25.389 ± 12.283 | 22.054 ± 8.331 | 0.080 |
| Liver function test (SGPT) | 17.551 ± 13.475 | 15.938 ± 11.705 | 0.475 |
| Liver function test (ALK) | 225.191 ± 80.826 | 208.482 ± 59.085 | 0.193 |
| Age, y | 45.904 ± 10.862 | 46.779 ± 11.840 | 0.06 |
| Addiction duration, mo | 12.518 ± 7.456 | 13.130 ± 9.290 | 0.638 |
| MMT duration, mo | 45.819 ± 24.126 | 40.500 ± 23.210 | 0.146 |
| Cigarettes per day (CPD) | 11.518 ± 9.257 | 10.453 ± 9.491 | 0.462 |
| Gender, No. (women: men: unknown) | 1:76:6 | 3:74:9 | 0.455 |
Abbreviations: HDD, high-dose dependent; LDD, low-dose dependent.
aValues are expressed as mean ± SD.
3.2. Genotyping
Blood samples were taken from all opioid-dependent subjects. DNA was extracted from the whole blood samples using a DNA extraction kit (Roche, Mannheim, Germany). ABCB1 SNPs at the C1236T, C3435T, A61G, G1199A, and G2677T loci were identified using polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP). The primer sequences used in this study for PCR and the size of the amplified fragments are shown in Table 2.
| Primer Name | Primer Sequence | Amplicon Size, bp |
|---|---|---|
| G1199A | 258 | |
| F* | 5 CAGCTATTCGAAGAGTGGGC 3 | |
| R | 5 CCGTGAGAAAAAAACTTCAAGG 3 | |
| A61G | 179 | |
| F* | 5 AGGAGCAAAGAAGAAGAACTTTTTTAAACTGATC 3 | |
| R | 5 GATTCCAAAGGCTAGCTTGC 3 | |
| C1236T | 527 | |
| F* | 5 CAGCTATTCGAAGAGTGGGC 3 | |
| R | 5 ATCCTGTCCATCAACACTGAC 3 | |
| C3435T | 207 | |
| F* | 5 TTGATGGCAAAGAAATAAAGC 3 | |
| R | 5 CTTACATTAGGCAGTGACTCG 3 | |
| G2677T | 224 | |
| F* | 5 TGCAGGCTATAGGTTCCAGG 3 | |
| R | 5 TTTAGTTTGACTCACCTTCCCG 3 |
3.3. PCR-RFLP
PCR-RFLP for all the assays was performed in a 20-mL mixture containing 100 ng extracted genomic DNA, 200 nmol/L of each primer, 200 mol/L dNTPs, and 1X PCR Master mix (Cinagen, Iran). The PCR was run on a thermal cycler in the following cycling conditions: 5 minutes at 95°C, 35 cycles of 30 seconds at 94°C, 40 seconds at 60°C, 1.5 minutes at 72°C, and a final extension step of 5 minutes at 72°C. A negative control reaction was run to avoid any contamination. Then, 2 µL of PCR products were run and visualized on 1% agarose gel electrophoresis.
The amplified fragments were digested using restriction endonuclease enzymes (Thermo Fisher Scientific, Massachusetts, USA): C1236T Eco 0109 (5U, 4 hours at 37°C and then 20 minutes at 65°C), C3435T Dpn II (5U, 16 hours at 37°C and then 20 minutes at 65°C), A61G Taq I (1U, 2 hours at 65°C), G1199A ACUI (2.5U, 16 hours at 37°C followed by 20 minutes at 65°C), and G2677T BanI (4U). After digestion, the restriction fragments were visualized on a 2% agarose gel electrophoresis, alongside a DNA molecular marker as a reference. The lengths of the restriction fragments were as follows: G1199A-wild-type, 206 + 52 bp and variant, 258 bp; A61G-wild-type, 179 bp and variant, 146 + 33 bp; C1236T-wild-type, 276 + 251 bp and variant, 527 bp; C3435T-wild-type, 145 + 62 bp and variant, 207 bp; and G2671T-wild-type, 198 + 26 bp and variant, 224 bp.
3.4. Data Analysis
The chi-square test was used to determine if the genotypes and alleles frequencies were in Hardy-Weinberg equilibrium. ABCB1 haplotypes of the two groups were obtained using the SNP analyzer version 2.0 software. The Fisher exact test and odds ratios were performed to determine any variability in the SNPs alleles, genotypes, and haplotype frequencies between LDD and HDD individuals. Linkage disequilibrium coefficients (D) among the variants for both groups were calculated using the SNP analyzer 2.0. The association of haplotypes and genotypes with daily methadone dose was tested with Kruskal-Wallis and Dunn multiple comparisons and ANOVA tests, respectively. Data are represented as means ± SD with a P value of < 0.05 as significant (Table 3).
| LDD Group (N = 86) | HDD Group (N = 83) | Total (N = 169) | Chi-Square | P Value | |
|---|---|---|---|---|---|
| C1236T | |||||
| Genotype | 0.7399 | 0.691 | |||
| CC | 23 (26.7) | 18 (21.7) | 41 (24.3) | ||
| CT | 38 (44.2) | 37 (44.6) | 75 (44.4) | ||
| TT | 25 (29.1) | 28 (33.7) | 53 (31.4) | ||
| Allele | 0.8026 | 0.370 | |||
| C | 84 (48.8) | 73 (44.0) | 157 (46.4) | ||
| T | 88 (51.2) | 93 (56.0) | 181 (53.6) | ||
| C3435T | |||||
| Genotype | 2.765 | 0.251 | |||
| CC | 26 (30.2) | 16 (19.3) | 42 (24.9) | ||
| CT | 23 (26.7) | 27 (32.5) | 50 (29.6) | ||
| TT | 37 (43.0) | 40 (48.2) | 77 (45.6) | ||
| Allele | 2.295 | 0.130 | |||
| C | 75 (43.6) | 59 (35.5) | 134 (39.6) | ||
| T | 97 (56.4) | 107 (64.5) | 204 (60.4) | ||
| A61G | |||||
| Genotype | 0.6197 | 0.431 | |||
| AA | 82 (95.3) | 81 (97.6) | 163 (96.4) | ||
| AG | 4 (4.7) | 2 (2.4) | 6 (3.6) | ||
| Allele | 0.6085 | 0.435 | |||
| A | 168 (97.7) | 164 (98.8) | 332 (98.2) | ||
| G | 4 (2.3) | 2 (1.2) | 6 (1.8) | ||
| G1199A | |||||
| Genotype | 0.2739 | 0.872 | |||
| AA | 3 (3.5) | 2 (2.4) | 5 (3.0) | ||
| GA | 7 (8.1) | 8 (9.6) | 15 (8.9) | ||
| GG | 76 (88.4) | 73 (88.0) | 149 (88.2) | ||
| Allele | 0.01337 | 0.908 | |||
| A | 13 (7.6) | 12 (7.2) | 25 (7.4) | ||
| G | 159 (92.4) | 154 (92.8) | 313 (92.6) | ||
| G2677T | |||||
| Genotype | 1.422 | 0.491 | |||
| GG | 25 (29.1) | 31 (37.3) | 56 (33.1) | ||
| GT | 38 (44.2) | 34 (41.0) | 72 (42.6) | ||
| TT | 23 (26.7) | 18 (21.7) | 41 (24.3) | ||
| Allele | 1.515 | 0.218 | |||
| G | 88 (51.2) | 96 (57.8) | 184 (54.4) | ||
| T | 84 (48.8) | 70 (42.2) | 154 (45.6) | ||
Abbreviations: HDD, high-dose dependent; LDD, low-dose dependent; SNP, single nucleotide polymorphisms.
aValues are expressed as No. (%).
4. Results
4.1. ABCB1 SNP Allele, Genotype, and Haplotype Frequencies
The frequencies of ABCB1 SNPs at each locus, regarding alleles and genotypes, showed no significant difference between the LDD and HDD groups and they were in Hardy-Weinberg equilibrium. In total, 11 haplotypes were observed in the groups by combining the individual ABCB1 SNPs into the haplotypes. The frequencies of haplotypes had no significant differences between the two groups except for haplotype 1 (P = 0.030557, χ2 = 4.678) as analyzed by the odds ratio and Fisher exact test performed for individual haplotypes (Table 4).
| ID | Haplotype | LDD Group | HDD Group | χ2 | P Value | OR | 95% CI |
|---|---|---|---|---|---|---|---|
| h1 | TTAGG | 55 | 72 | 4.678 | 0.030557 | 1.629 | 1.045 - 2.539 |
| h2 | CCAGT | 40 | 35 | 0.231 | 0.631006 | 0.882 | 0.527 - 1.474 |
| h3 | CTAGT | 16 | 15 | 0.007 | 0.93245 | 0.969 | 0.463 - 2.028 |
| h4 | CCAGG | 11 | 7 | 0.795 | 0.372553 | 0.644 | 0.244 - 1.704 |
| h5 | TTAGT | 10 | 6 | 0.906 | 0.341125 | 0.608 | 0.216 - 1.711 |
| h6 | TCAGG | 8 | 7 | 0.038 | 0.846311 | 0.903 | 0.32 - 2.547 |
| h7 | CTAGG | 7 | 7 | 0.005 | 0.945905 | 1.038 | 0.356 - 3.026 |
| h8 | TCAGT | 8 | 4 | 1.239 | 0.26557 | 0.506 | 0.149 - 1.714 |
| h9 | CCAAT | 3 | 3 | 0.136 | 0.712795 | 1.037 | 0.206 - 5.212 |
| h10 | TTAAT | 1 | 3 | 0.29 | 0.590035 | 3.147 | 0.324 - 30.566 |
| h11 | CTAAG | 2 | 2 | 0.218 | 0.640254 | 1.037 | 0.144 - 7.446 |
Abbreviations: CI, confidence interval; HDD, high-dose dependent; LDD, low-dose dependent; OR, odds ratio.
4.2. Linkage Equilibrium
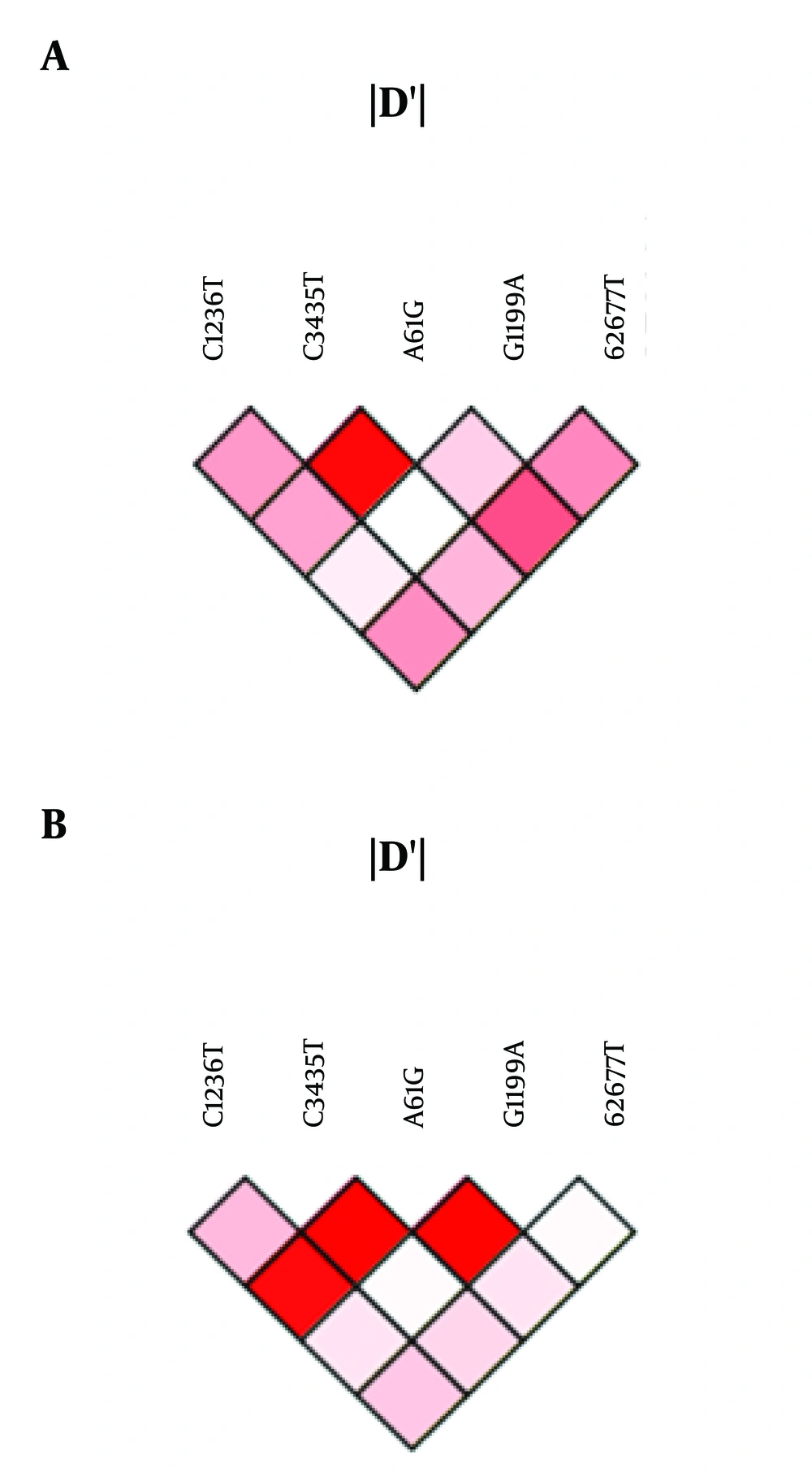
Linkage disequilibrium analysis was performed using the SNP analyzer 2.0. In the HDD group, we observed linkage between C3435T and A61G, A61G and G2677T, G1199A and G2677T, C1236T and G2677T, and C1236T and A61G (Figure 1A). Moreover, a strong linkage was observed between C1236T and C3435T, C3435T and A61G, and A61G and G1199A in the LDD group (Figure 1B).
4.3. Relationships of ABCB1 Genotype and Haplotype with Methadone Dosage Requirements
The average methadone doses were 127 ± 2.33 and 42.14 ± 12.74 in the HDD and LDD groups, respectively. In order to see the effect of haplotypes on the methadone dosage requirement, we analyzed the methadone dose in the HDD and LDD groups carrying h1 haplotype with a significant difference in haplotype frequency between the two groups. However, the methadone dose received by the h1 haplotype carriers was not significantly different from the dose received by the h1 non-carriers in the HDD group (126.95 ± 32.36 vs. 127.16 ± 22.77, respectively; P = 0.8922). Moreover, no significant difference in the methadone dose was observed between h1 carriers and non-carriers in the LDD group (41.72 ± 13.10 vs. 42.44 ± 12.58, respectively; P = 0.88). In addition, we compared the methadone dose requirement between different haplotypes in the whole sample; however, no significant difference was detected by pairwise comparison of the haplotypes (Table 4).
To test the effect of different SNPs at different loci on the methadone dosage requirement, we compared the mean methadone dose between different genotypes in the whole sample using the ANOVA test. None of the SNPs showed significant differences in the daily methadone dose between different genotypes. The mean methadone doses were 79.5 ± 46.86, 83.47 ± 47.99, and 87.64 ± 48.87 at C1236T locus for CC, CT, and TT genotypes, respectively (P = 0.71), 73.41 ± 47.55, 87.94 ± 43.87, and 86.68 ± 50.2 at C3435T locus for CC, CT, and TT, respectively (P = 0.27), 84.38 ±4 8.2 and 69.17 ± 35.84 at A61G locus for AA and AG, respectively (P = 0.44), 82.88 ± 47.51, 92 ± 50.31, and 88 ± 57.73 at G1199A locus for GG, GA, and AA, respectively (P = 0.76), and 89.91 ± 45.96, 81.79 ± 50.8, and 79.15 ± 45.12 at G2677T locus for GG, GT, and TT, respectively (P = 0.49).
5. Discussion
It is estimated that there are approximately 16 million opioid-dependent people worldwide, a situation related to the intense morbidity and mortality from overdose and infections such as hepatitis, tuberculosis, and HIV (17). Illicit drug use imposes social costs in terms of public health, crime, and productivity. Methadone has been approved by the WHO as the main maintenance treatment for opioid dependence (7). It dramatically decreases HIV and hepatitis infections, crimes, and overdose mortality (18). However, due to the high genetic variability among individuals, personalized dosing is required to achieve the optimal outcomes for methadone treatment.
P-glycoprotein, a protein encoded by the ABCB1 gene, transports methadone at the blood-brain barrier (BBB) in the CNS. Genetic polymorphisms in this gene may affect the expression or functional activity of P-glycoprotein resulting in the methadone uptake in the CNS (19). We aimed to investigate the relationship between the ABCB1 gene variability and methadone dose requirement in Iranian opioid-dependent people.
The five most frequently reported ABCB1 SNPs (C1236T, C3435T, A61G, G1199A, and G2677T) (12) were selected and included in our study for investigation. The C3435T SNP showed the highest frequency in both LDD and HDD populations (43.0% and 48.2%, respectively). However, in line with previous studies on white populations (20, 21), we did not find any significant difference in the allele and genotype frequencies between HDD and LDD subjects.
In addition, haplotype analysis showed a combination of 19 ABCB1 haplotypes for these five SNPs, of which 11 were unique. The results showed that only had the TTAGG haplotype a significantly higher frequency in the HDD population than in the LDD group (86.74% vs. 63.95%, respectively; P = 0.030557, χ2 = 4.678), indicating a higher risk (OR = 1.6, 95% CI = 1.045 - 2.539) of methadone use in HDD subjects. There was no significant difference in other haplotype frequencies between the two populations. These results underlie no link between the variability in the ABCB1 gene and drug dependence. Previous reports have shown linkage disequilibrium between C1236T, G2677T, and C3435T SNPs (15, 22, 23). Our linkage analysis results showed a strong linkage at the 3435, 61, and 2677 loci in the HDD population and at 1236, 3435, 61, and 1199 loci in LDD subjects. However, no variant was observed at the 61 locus indicating that its linkage is unlikely to be real.
There are only a few reports of the frequency of ABCB1 haplotypes in different white populations. However, the haplotype analysis results in our study are comparable to the reports of previous studies. Kim et al. (23) previously reported that the C1236T and C3435T SNPs were linked and occurred with a frequency of 62% in healthy European Americans and 13% in healthy African Americans. They reported a frequency of 35% for the wild-type haplotype. In addition, an intermediate frequency of 15% (16 from abc) and 28.3% - 30.8% (Abc ref) for the wild-type haplotype was previously observed in different white populations. In addition, previous studies have found a higher frequency for AGTTT variant haplotype (25.8% - 35% (24), 32% (15), and 27% (23)) than our study (7.22% - 11.62%). We suggest the differences in haplotype frequencies could be probably due to the number of SNPs investigated in previous studies. Of note, in our study, the wild-type haplotype carriers were more than variant haplotype carriers while in previous studies, fewer individuals carried the wild-type haplotypes. Furthermore, the observed differences in the haplotypes may also be due to the different ethnicity of the study populations.
Moreover, the possible impact of the ABCB1 haplotypes on the methadone dose requirement was tested. However, no significant difference was detected in the methadone dose requirement between haplotype 1 carriers and haplotype 1 non-carriers in both populations. In addition, the investigation of the ABCB1 genotypes on the methadone dose received by the whole population identified no significant difference between different genotypes of the SNPs. Consistent with our findings, Lotsch et al. reported that G2677T and C3435T variants had no effect on the drug uptake and distribution in healthy individuals (25).
In contrast, Coller et al. found that subjects carrying two copies of the wild-type haplotype received a higher dose of methadone than those with only one or no copy of wild-type haplotype (24). In comparison, first, their study included a smaller number of subjects than our study, which could influence the frequency of haplotypes in the population. Second, their study was conducted in a population of opioid-dependent subjects and healthy controls while our study included HDD and LDD subjects. In addition, because we recruited our subjects from the Iranian population, ethnicity may be a possible factor in making differences. Another study conducted by Wang et al. demonstrated that the C3435T variant affected mRNA stability and decreased P-glycoprotein expression leading to a lower methadone dose requirement (26). However, we did not find any difference in methadone dose requirement between subjects carrying the C3435T variant and non-carriers. This could be due to the small number of subjects in our study.
Although our study is limited by the number of subjects, this is the first report of genetic variability of ABCB1 in Iranian opioid-dependent individuals. However, further studies with larger sample sizes are required to achieve a more accurate frequency of the reported SNPs and their impact on daily drug use.
5.1. Conclusions
In conclusion, we identified eleven unique haplotypes of ABCB1 SNPs in HDD and LDD subjects. We observed a strong linkage between the SNPs in the two populations. Thus, it seems that the use of these SNPs to predict and optimize the amount of drug in Iranian populations is not beneficial. However, to gain a better understanding of the impact of genetic variability on the optimal drug dose requirement, large-scale haplotyping studies are required that could provide optimal treatment and decrease social costs.