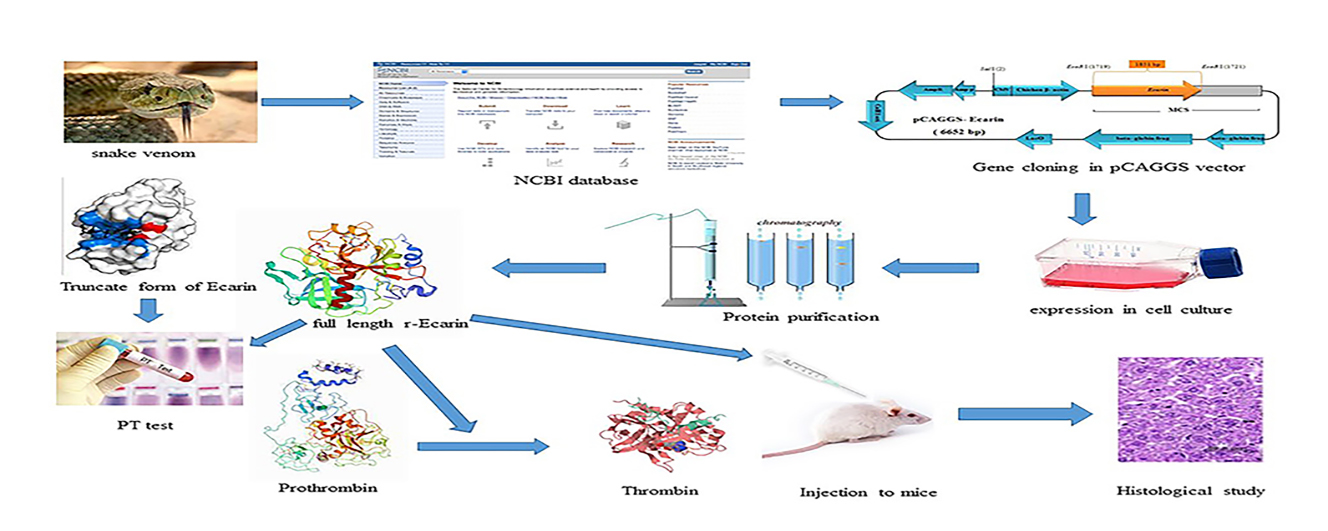
1. Background
Quantification of functional and dysfunctional prothrombin is an important clinical requirement. Functional prothrombin is a vitamin K-dependent protein with a molecular weight of 72 kDa organized into four domains: an N-terminal Gla domain, two kringle domains, and a C-terminal protease domain. The cleavage of Gla and kringle sites in a Ca2+-dependent process by Xa and Va gives rise to the formation of intermediates such as prethrombin-2 or meizothrombin (1). The GLA domain is composed of 10 glutamate residues, and after translation by vitamin K-dependent carboxylation, it is converted into γ-carboxyglutamate (Gla). In coagulation proteins, all glutamic acid (Glu) residues are converted into Gla (2). Nonfunctional prothrombin or Des-γ-carboxy prothrombin (DCP) does not have the GLA domain. In HCC (hepatocellular carcinoma) patients, those with vitamin K deficiency, and those treated with warfarin, there is a nonfunctional form of prothrombin present. The abnormal prothrombin cannot be activated by factor Xa and, thus, these patients have coagulation disorder (3). In the blood, thrombin is produced only via proteolysis of normal zymogen prothrombin by the prothrombinase complex (prothrombin activator), consisting of Xa, Va, Ca2+, and phospholipid (4). Prothrombin activators are also found in snake venom (5, 6). Ecarin is a P-III snake venom metalloprotease (SVMP) that includes three domains, metalloproteinase (M), disintegrin-like domain (D), Cys-rich domain (C), and has 64% identity with the H chain of RVV-X (Russell's viper venom X activator) (7). This enzyme affects both functional and abnormal prothrombin by cleaving the 320-Arg-Ile-321 bond and converts them into meizothrombin or prethrombin-2, which, subsequently, is converted into α-thrombin by autolysis (8). Unlike other prothrombin activators in snake venom, Ecarin does not require cofactors, phospholipids, Ca2+, or the presence of GLA domain for its protease activity (9-11). Since the isolation of prothrombin activators from snake venom has many drawbacks and importing them to the country costs a lot, production of recombinant enzymes (such as r-Ecarin) in the eukaryotic expression systems such as CHO cells is a good alternative for these native compounds. With this method, we can stably express large quantities of r-Ecarin for use in a laboratory test to evaluate anticoagulation during treatment with synthetic thrombin inhibitors such as melagatran, argatroban, and hirudin (12, 13). It can also be used for determining total prothrombin in plasma or buffer, or, in other words, can be used for tracing low serum prothrombin levels, for studying coagulation disorders, and for quantitative measurement of abnormal prothrombin in patients with hepatic disorders (such as hepatocellular carcinoma) or those with vitamin K deficiency. Ecarin can also trace lupus anticoagulants levels in the patient’s serum (14, 15). Meizothrombin as an intermediate in the thrombin production pathway of Ecarin can act as a neutralizer of serum toxic hirudin levels, so Ecarin can be used in anticoagulant therapy with hirudin (16-18). The main problem in low specificity diagnostic tests is its contamination and high cost, which will cause a non-specific response of the enzyme used against the substrates. Therefore, any diagnostic and therapeutic test requires an enzyme or part of its structure so that it has sufficient sensitivity and specificity. Conventional coagulation tests are not able to detect anticoagulants quickly and accurately, but Ecarin enzyme in Ecarin clotting time (ECT) and Ecarin chromogenic assay (ECA) can detect these anticoagulants quickly and specifically. Therefore, it can use to evaluate the treatment of many diseases such as cancer, liver, and cardiovascular diseases. Early diagnosis can lead to significant advances in the control and treatment of diseases.
2. Objectives
The aim of this study was to produce safe, cost-effective, and high-performance r-Ecarin for diagnostic, and therapeutic purposes. Comparing the expression and function of full-length and truncated forms of this protein obtained from the previous study (19) helps to select the right one for production.
3. Methods
3.1. Docking of Ecarin and the Substrate Prothrombin
The Swiss-model server (https://swissmodel.expasy.org), a SIB Bioinformatics Resource Portal for protein structure and function characterization, was used to make the Ecarin homology model (20). Using 2e3x.1.A (crystal structure of Russell's viper venom metalloproteinase with 66.02%. identity) as a template (21), the I-TASSER program (http://zhang.bioinformatics.ku.edu/I-TASSER) was used to predict the three-dimensional structure of the protein (22). The ConSeq program (http://consurf.tau.ac.il/) was used to analyze the evolutionary conservation of amino acids of the protein (23). One of the target proteins for Ecarin is prothrombin with 2hpq PDB code. The ClusPro web server (https://cluspro.bu.edu/login.php) was used for docking analysis (24). Then, using the PDBsum server, we have provided an at-a-glance overview of each three-dimensional structure placed in the Protein Data Bank (http://www.rcsb.org/pdb/home/.do).
3.2. Construction of Plasmid
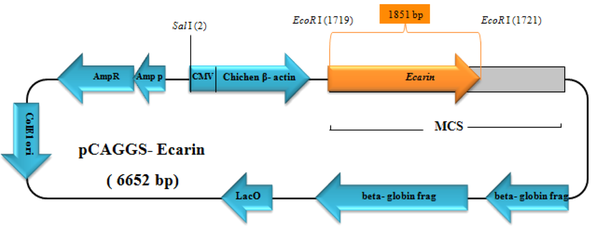
An 1851-bp nucleotide sequence of the Ecarin gene was obtained from the NCBI (accession number: D32212), and then it was artificially synthesized (GenCust, Luxembourg) into a pBluescript vector. After transforming the vector into TOP10 Escherichia coli, it was cultured on ampicillin-LB agar. To subclone the Ecarin gene into an expression vector, it was digested with EcoRI (Fermentas, Lithuania) restriction enzyme. The product of digestion was analyzed by electrophoresis on 0.8% agarose gel, and the target band was removed and purified using a DNA gel extraction kit (Qiagen, USA). The purified Ecarin was inserted into a digested pCAGGS expression vector, and then the ligation mixture transformed into E. coli (Figure 1). Recombinant plasmid extraction was performed using a plasmid extraction kit (Qiagene, USA). The recombinant plasmid was confirmed using polymerase chain reaction (PCR), and specific primers of the Ecarin included forward (5´GACCCTGGGCGAATTCGTGCCACC3´) and reverse (5´ATGCAATGTCGAATTCGAGAGGTG3´) primers and digestion by EcoRI restriction enzyme.
3.3. The CHO- Ecarin Stable Line and Expression of the Ecarin Protein
The Chinese Hamster Ovary cell line (CHO; DG44 was a gift by Dr. Mahboudi) was routinely cultured under sterile conditions in the DMEM culture medium containing 10% fetal calf serum (FCS). pCAGGS-Ecarin was transfected into the CHO cell using electroporation (350 V, 500 μs). Transfected cells were then seeded in six-well culture plates with DMEM containing G418 (Santa Cruz, USA) (100 μg/mL) and incubated at 37°C with 90% humidity under 5% CO2 conditions for approximately 14 days. Controls, including CHO cells, transfected with empty pCAGGS and untransfected CHO cells were used in this study.
3.4. Purification and Confirmation of the Ecarin Protein
For the purification of Ecarin protein, a Macro-Prep High S Support (20 mL. Bio-Rad Laboratories) column was washed with 20 mM citrate (pH 5.0) buffer. The pH of the culture medium was adjusted to 5.0 using citric acid (1 M) and then applied to the column. The column was washed with washing buffer and then eluted with a salt-concentration gradient (100 - 700 mM NaCl/20 mM citric acid). Fractions were used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis with anti-Ecarin antibody (25).
3.5. The Function of Recombinant Ecarin
To determine the activity of the produced r-Ecarin, Tris buffer consisting of 100 mM NaCl and 20 mM Tris HCl, with pH 8, was added to prothrombin (1 mg/mL) and 50 mM benzamidine. The r-Ecarin (2 U/mL) was then added to the reaction and incubated for 10 minutes at 37°C (25). Then, the generated thrombin and Ecarin expression in transfected cells were evaluated by SDS-PAGE.
3.6. Measurement of Protein Coagulation Activity
One-hundred microliter of citrated plasma (ratio 1.9) was added to the tube and incubated for 2 min at 37°C. We then added 200 µL thromboplastin (vials containing thromboplastin and calcium) to the plasma tube and the blood clotting time was recorded immediately using a stopwatch. The normal time for this test is 12 - 14 s. Then 0.5 mg/mL concentration was prepared from both r-Ecarin and r-Ecamet proteins. This concentration was tested by PT to evaluate the coagulation properties of proteins.
3.7. SDS PAGE and Western Blot Analysis
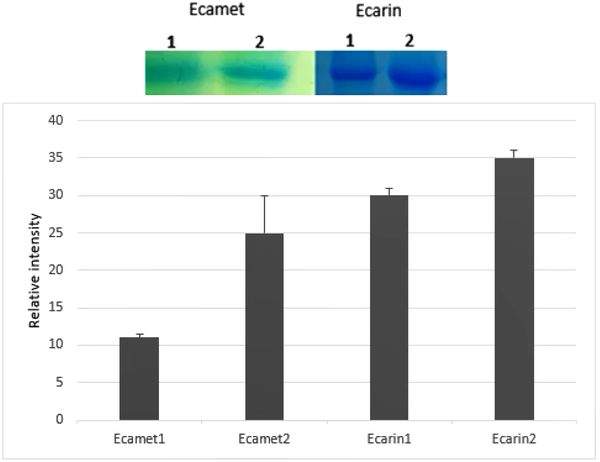
The supernatant of the transfected cells was precipitated with TCA (Trichloroacetic acid) then mixed with 2 x SDS-loading buffer (20% glycerol, 4% SDS, 0.005% bromophenol blue, 100 mM Tris-HCl pH 6.8, and 200 mM DTT) and heated at 85°C for 5 min and applied for gel electrophoresis (25). SDS-PAGE was performed using a 12% (w/v) polyacrylamide gel according to the standard protocol. The gel was stained with Coomassie blue G250 and then destained. Commercial Ecarin (Merck, 250 μg) was injected into the mouse with Freund complete adjuvant on days 0, 14, and 28 to produce a polyclonal antibody against the Ecarin, and then western blot analysis was used to confirm the presence of antibodies against Ecarin. For western blotting, the gel-separated proteins were transferred to the 0.2 μm nitrocellulose membrane (Sigma-Aldrich) using a mini trans-blot electrophoretic transfer cell (Bio-Rad, USA). The membrane was then blocked with 3% skimmed milk powder in phosphate-buffered saline (PBS) for 1 h. After washing the membrane three times using Tris-buffered saline with Tween 20 (TBST) (0.5 M NaCl, 0.02 M Tris pH 8.5, 0.05% Tween 20), the membrane was incubated with prepared polyclonal anti-Ecarin antibodies for 2 h at 37°C, followed by washing as previously described. The membrane was then incubated with 1:10 000 diluted goat anti-mouse immunoglobulin alkaline phosphatase (ALP) conjugate at a shaking speed of 50 rpm for 2 h. After washing, the corresponding protein bands were detected with 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitroblue tetrazolium (NBT). The expression level of r-Ecarin quantified by SDS-PAGE method and Image J software (http://rsb.info.nih.gov/ij) was compared with the expression of r-Ecamet protein.
3.8. Evaluation of Cytotoxicity Effect of r-Ecarin and r-Ecamet Proteins on the Liver
This study was performed on three female Balb/c mice. These animals were purchased from Pasteur Institute (Tehran, Iran), and to prepare for the experiment, they were kept for two weeks in the animal room in special cages at a suitable humidity of 50 - 70°C and a suitable laboratory temperature (23 ± 2°C). Then, 200 (μg/mL) recombinant protein was prepared from each of the proteins (Ecarin and Ecamet) and injected intraperitoneally into mice under sterile conditions. A mouse as control was injected with phosphate-buffered saline (PBS). Then, the mice’s livers were prepared for histopathological analysis.
4. Results
4.1. Evaluation of the Docking Between the Homology Models
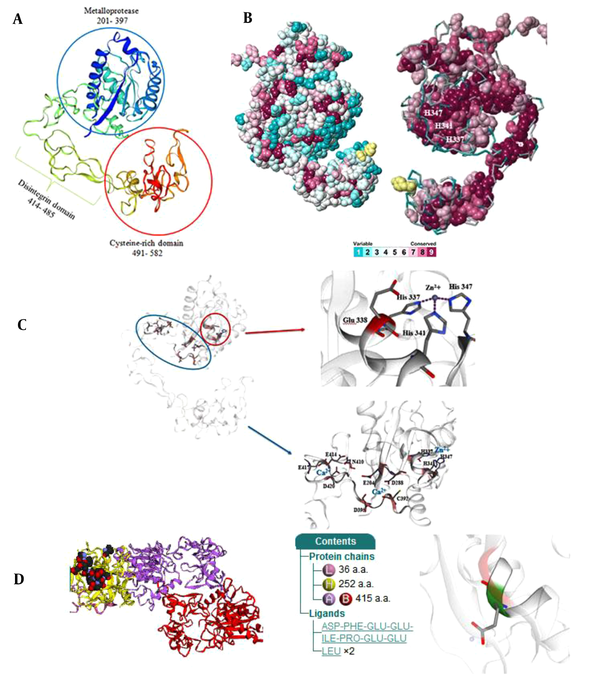
The three-dimensional structure of Ecarin was modeled using Chain A of Russell’s viper venom metalloproteinase as the template with 66.02% identity (Figure 2A). In the active site pocket, conserved residues, H337, H341, H347, and E338, are required to bind Zn2+ and E204, D288, D395, C392, N410, E414, E417, and D420 are required as binding sites for Ca2+ cofactor (Figures 2B and 2C). Molecular docking studies of prothrombin, a blood procoagulant factor, and Ecarin, a metalloproteinase, were performed using the modeled protein structure of Ecarin. In the first cluster, it contains 94 members with the lowest energy score of –323.2, which means that in the conformational space, the target complex has a minimum of Gibbs free energy. Figure 2D shows the functional site and the best format of the produced complex between two proteins. It also shows the molecules that construct the structure and schematic diagrams of the interactions between them with a chain containing ASP 355 to GLU 362 (ASP-PHE-GLU-GLU-ILE-PRO-GLU-GLU) from Ecarin.
Three-dimensional models, Zn2+-binding pocket, Ca2+ binding sites, and conserved residue for Ecarin. (A) Cartoon view of the 3D structure of Ecarin. The three main domains: M, D, and C, are shown. (B) Conserved residue analysis of Ecarin. Variable to highly conserved residues are shown in blue to red, respectively. (C) The active site and Zn2+-binding pocket, the Ca2+ binding residues are demonstrated (Color figure online). (D) Schematic view of complex formation Ecarin and Prothrombin. PDB id: Y925 shows amino acids from Ecarin in interaction with prothrombin.
4.2. Preparation and Cloning of Ecarin Gene
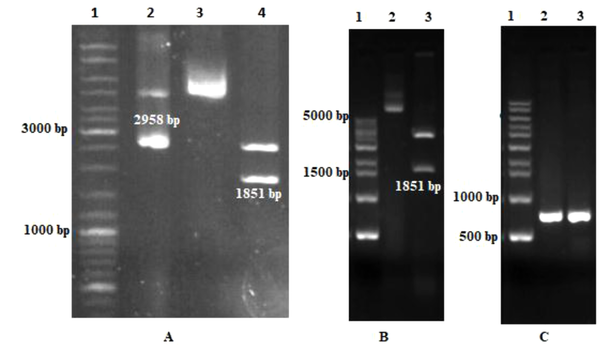
The synthetic plasmid (pBluescript-Ecarin) was constructed and transformed into top 10 E. coli bacteria and cultured on ampicillin-LB agar. pCAGGS vector was used as an expression vector for Ecarin subcloning. Digestion of pBluescript containing Ecarin insert (Figure 3A) and pCAGGS plasmid and then gel electrophoresis caused the isolation of Ecarin gene (1851 bp) and a linear pCAGGS (4800 bp). Ligation of Ecarin and linear pCAGGS led to the construction of recombinant expressional vector. Then, the vector was transformed into E. coli cells and grown on the LB agar plate with ampicillin. To confirm the cloning of expression pCAGGS, plasmids of three clones obtained from the ligation reaction were extracted and digested with EcoRI (Figure 3B). For further validation, recombinant pCAGGS was confirmed using PCR through specific primers of the metalloprotease domain of Ecarin gene. These primers produced a product of 655 bp (Figure 3C).
Gel electrophoresis of the recombinant plasmid containing Ecarin gene. (A) Digestion of pBluescript vector containing Ecarin fragment by EcoRI enzyme. Lane 1: 1 kb DNA ladder; Lane 2: pBluescript vector as a control; Lane 3: Recombinant pBluescript; Lane 4: digested synthetic recombinant pBluescript with EcoRI. (B) Confirmation of recombinant pCAGGS with digestion. Lane 1: DNA ladder. Lane 2: Recombinant pCAGGS vector as a control; Lane 3: digested pCAGGS vector with EcoRI. (C) Confirmation of recombinant pCAGGS with PCR. Lane 1: DNA ladder; Lane 2 and 3: amplification of the Ecarin gene with specific primers of the gene (655 bp).
4.3. Expression Analysis and Purification of r-Ecarin Protein
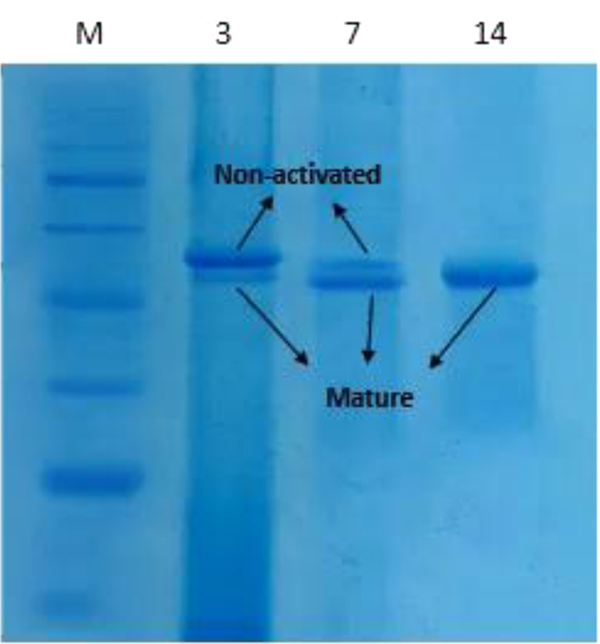
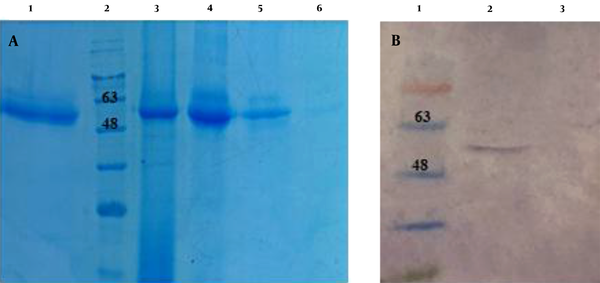
In order to analyze the Ecarin protein expression, recombinant pCAGGS-Ecarin was transfected into CHO cells. Then, protein expression was assessed and compared with the control CHO cells using SDS-PAGE. Ecarin is produced as a pro-protein, and removal of pro-peptide from Ecarin causes its optimal activity. Our results showed that by incubating the culture medium for 14 days, mature Ecarin is entirely produced. Figure 4 shows that after three days of incubation of the culture medium there is a large amount of immature Ecarin, which is greatly reduced on the seventh day and mature Ecarin reaches its maximum by the 14th day after complete cell death. The expressed protein showed a molecular weight of 55 kDa, which is not present in controls. Purification of the protein was performed by ion-exchange chromatography CM-cellulose according to the preparation protocol (25). Elution buffers with a pH gradient were used to wash the resin column, and then SDS analysis was performed for each elution (Figure 5A). The result of the expressed band was confirmed by western blotting (Figure 5B). Quantitative Expression Analysis of full-length and truncated recombinant Ecarin displayed a high degree of differential expression in these proteins. Full-length recombinant Ecarin showed a higher expression level (Figure 6).
(A) SDS-PAGE of Ecarin protein. Lane 1: commercial Ecarin as control; Lane 2: protein size marker; Lanes 3-6: Ecarin purified fragment. (B) Confirmation of the expressed Ecarin by Western blot analysis. Lane 1 protein size marker; Lane 2: transfected cells containing pCAGGS- Ecarin; Lane 3: negative control (pCAGGS without Ecarin fragment).
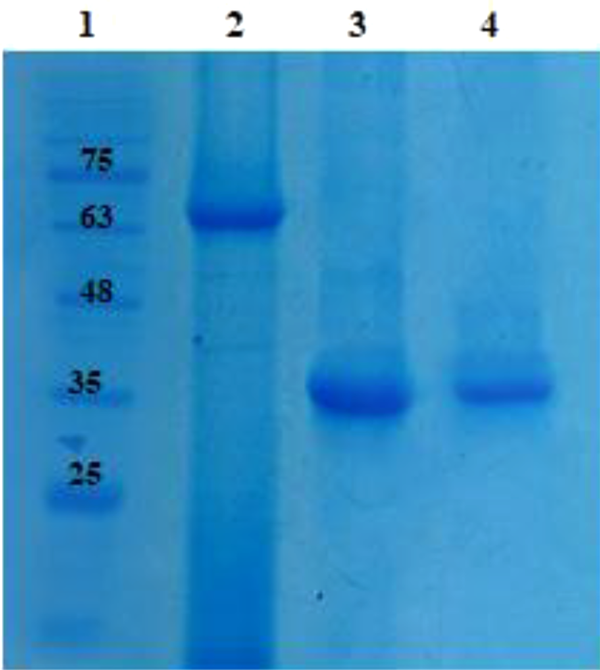
4.4. Conversion of Prothrombin to Thrombin by r-Ecarin
For bioactivity analysis of protein, a chemical reaction was performed (in accordance with the protocol mentioned in the methods). Results of this assay showed that r- Ecarin can digest prothrombin (approximately 72 kDa) and produce thrombin with a molecular weight of approximately 35 kDa (Figure 7). Due to the clotting time of normal (protein-free) plasma, it was found that r-Ecarin and r-Ecamet proteins clots the plasma rapidly. The clot formed quickly by adding protein at a concentration of 0.5 mg/mL, so these proteins have a coagulation property. The obtained times for PT test with three replications for these proteins were analyzed using SPSS software version 23 and T-Test statistical test. There was a statistically significant difference in the activity of the two enzymes of Ecarin (P < 0.05). The activity time of r-Ecarin was 5.33 ± 15 Second (Mean ± SD) and the activity time of r-Ecamet was 27.6 ± 2.51 Second (Mean ± SD). This comparison showed that full-length protein has a more significant coagulation effect than truncated form (Table 1).
| Protein Concentration | PT (s) Mean | Description |
|---|---|---|
| 0.5 (mg/mL) r-Ecamet | 27.6 ± 2.51 | The clot is fully formed. |
| 0.5 (mg/mL) r-Ecarin | 5.33 ± 15 | The clot is fully formed. |
| Normal PT (without protein) | 3.75 ± 30 | The clot is fully formed. |
PT Test for 0.5 mg/mL Concentration of r-Ecamet and r-Ecarin Proteins
4.5. Histopathological Analysis
In microscopical evaluation, in all three cases (control and tested cases), no obvious pathological changes related to the treatment, and no noticeable difference were detected between the tested and control groups. No architectural distortion was detected in cases (Figure 8).
5. Discussion
Numerous biochemical processes lead to the formation of blood clots. A wide range of changes can affect coagulation factors and lead to some congenital and acquired abnormalities. The studies showed that there is a lack of coagulation proteins production in hemophilia A and B, Von Willebrand, lupus, cardiovascular disease, and liver dysfunction or vitamin K deficiency (26). Animal toxins and secretions are a rich and complex combination of biologically active proteins and peptides that have shown many functions on the hemostatic system. Due to the biochemical and pharmacological properties of these toxins, they can be used in the diagnosis, treatment, and follow-up of many diseases related to disorders of the hemostatic system (27). Snake venom is a source of metalloproteases and serine proteases that affect blood clotting, fibrin lysis, and the complement system. One of these metalloproteases is Ecarin, which plays an important role in blood coagulation (28). The purpose of this study was to produce a biologically active form of r-Ecarin through a synthetic construct and comparison its expression and function with the truncated form of it contains active site fragment. Evaluation of purified proteins expression and function has shown that there is a significant difference between the two recombinant proteins, and r-Ecarin protein has more expression and better enzymatic function than truncated form. It can convert prothrombin into thrombin and can be a good alternative to natural Ecarin in diagnostic tests. In other investigations, a researcher named Kornalik et al. first isolated a calcium-independent coagulation protein from the saw-scaled viper Echis carinatus, which was initially named prothrombin-activating principle and later changed to Ecarin (29). Initially, Ecarin was introduced as a fibrinogenolytic protein, but after complete purification, it was found that, unlike other venom metalloproteases, it has no fibrinogenolytic activity and shows high specificity for prothrombin (30). Another researcher, Nishida et al. cloned Ecarin cDNA from the venom glands of the Kenyan snake Echis pyramidum leakyi and identified its amino acid sequence. Their results showed that the His-Glu-Xaa-Xaa-His-Xaa-Xaa-Gly-Xga-Xaa-His sequence in the metalloproteinase domain of Ecarin is the binding site of the Zn2+. This protein has four to five N-glycosylation sites that play a role in its solubility (7). Also, due to the presence of 35 cysteine amino acids in Ecarin sequence, this protein has high stability, and the instability index (II) was estimated at 39.25 (11). Our results showed that r-Ecarin remained stable for 14 days in culture medium at room temperature. For this protein, a single-chain glycoprotein, different molecular weights from 55 to 86 kDa have been reported, which may be due to differences in post-translational changes, including glycosylation, or the diverse subspecies of snakes considered (10, 31). In our study, the molecular weight of r-Ecarin was 55 kDa, which was consistent with previous results. In the other study, Jonebring et al. compared the function of recombinant and native Ecarin. They showed that r-Ecarin converts prothrombin into thrombin faster than native Ecarin and has higher activity at higher prothrombin concentrations (32). Numerous studies on this enzyme have led to the development of ECT and ECA tests (33-35). Using these tests, the consumption of thrombin inhibitors such as heparin and hirudin in various diseases is controlled to prevent severe bleeding (36). This protein affects both prothrombin and acarboxyprothrombin by cleaving the 320-Arg-Ile-321 bond and converting them into meizothrombin, which, subsequently, is converted into α-thrombin by autolysis (37). The enzyme can also affect bovine thrombin, which undergoes mutation in the active site and cleaves it at the Arg323-Ile324 and meizothrombin autolysis site. The optimum pH for Ecarin protease activity is 8 - 8.5, which is inhibited by chelating agents (1, 10-phenanthroline, EDTA), alkylating agents (iodoacetamide), and sulfhydryl blocking agents (L-cysteine, ascorbate, 2-ME, glutathione, DTT) (30, 38). The biological activity of Ecarin can be assessed by measuring the coagulation times of citrated human plasma or by measuring the meizothrombin activation using a chromonic substrate. Stocker et al. used tosyl-glycyl-L-prolyl-L-arginine-pnitroanilide (Chromozym TH) chromogenic substrate to measure the prothrombin converting potency of Ecarin because it has no inhibitory or activating effect on prothrombin conversion (39). Studies showed that the disintegrin domain increases the metalloproteinase activity of the enzyme and the presence of cysteine-rich and disintegrin domains contribute to the formation of the Ecarin c-shaped conformation, in which case cysteine-rich domain approaches the metalloproteinase domain. C-shaped conformation causes flexibility between the active site and the exosite of the enzyme and plays an important role in the identification of the substrate (40). Therefore, these two domains have a significant effect on proper folding and the biological activity of snake venom metalloproteases (41). This is in line with our findings, which show that full-length Ecarin has a higher activity.
5.1. Conclusions
Expression and purification of r-Ecarin provide a simple, rapid, and cost-effective procedure for producing a standardized activator of prothrombin. We have successfully produced r-Ecarin in CHO cells. Comparison of r-Ecarin function and expression with r-Ecamet as a truncated form of protein containing metalloprotease domain showed a significant difference between two proteins. r-Ecarin can prove to be a suitable alternative for native, synthetic, or even truncated forms of Ecarin in laboratory tests and clinical studies.