1. Background
Pentagamavunon-1 (PGV-1) 2,5-bis-(4-hydroxy, 3’,5’-dimethyl)-benzylidine-cyclopentanone is a curcumin analogue that exhibits anti-cancer properties toward several cancer cells (1). Further study reveals that PGV-1 displays a stronger cytotoxic effect than curcumin toward 4T1 breast cancer through various effects: G2/M arrest induction, cell migration inhibition, cellular senescence induction, and intracellular reactive oxygen species (ROS) level generation (2). PGV-1 prevents some ROS-metabolic enzymes and suppresses tumor formation in vivo of K562 bearing mice but shows low adverse effects that may correlate to the pre-metaphase arrest but remain unknown to the particular target proteins (3). The potential tumor suppressing agent of PGV-1 is also demonstrated in an in vivo model employing 4T1 cells, showing that PGV-1 exhibits a more powerful anti-cancer effect against triple-negative breast cancer (TNBC) (4T1) tumors development than curcumin did (4). Accordingly, with the relatively low IC50 value against 4T1 cells, PGV-1 is promising to be applied as a chemotherapeutic agent for TNBC. However, the effectiveness of PGV-1 remains to be increased to achieve a lower concentration of its cytotoxic effect. In addition, the mechanism of mitotic arrest is necessary to be explored further.
Adjunctive therapy with natural products is commonly applied to enhance the cytotoxic effect or evade the adverse effects of chemotherapeutic agents. Piperine, the main compound of Piper nigrum L., is interesting to offer the chance in combinatorial treatment with PGV-1 due to its pharmacological characteristic to increase drug bioavailability. The addition of piperine to the therapeutic agent increases the effectiveness of the drug (5). Piperine increases curcumin's anticancer activity against HCT116 cells (6). Interestingly, curcumin and piperine also suppress P-gp expression and function (7). P-gp (P-glycoprotein 1), also known as multidrug resistance protein 1 (MDR1), is a crucial cell membrane protein involved in the exit of many foreign compounds (7). Considering that PGV-1 has structural similarities to curcumin, the combination of PGV-1 and piperine may enhance the cytotoxic effect of PGV-1 synergistically. Additionally, piperine suppresses cell proliferation and causes 4T1 cells to enter the G2/M phase of the cell cycle (8), while PGV-1 promotes mitosis arrest specifically at pre-metaphase (3). Therefore, targeting mitotic arrest is a promising strategy of an agent against cancer cells.
Mitosis is mainly regulated by CDK1 (cyclin-dependent kinase 1) that is activated by binding to cyclin B1 (9). If DNA damage is detected, CHK1 (checkpoint kinase 1) prevents the entry of mitosis through inhibition of CDK1 activity through the decrease of cyclin B1expression (9), leading to the accumulation of cells at the G2 phase temporarily (10). Nevertheless, because cancer cells' G2 checkpoints are damaged mainly, they are unable to maintain G2 arrest and eventually perish when they enter mitosis (10). This process is known as mitotic catastrophe characterized by the formation of giant cells with polynuclear (11, 12). As most cells that go through mitotic catastrophe die, cellular mechanisms that cause irreversible growth arrest are referred to as "reproductive death" or categorized as senescence (13). It is in line with previous findings, which revealed that PGV-1 induces G2/M arrest against TNBC cells correlates with senescence evidence (2). Furthermore, the target proteins for piperine and PGV-1 for TNBC co-treatment need to be explored.
In TNBC cells, it is known that the majority of regulator genes in the cell cycle are upregulated, including CDK1 (14). Therefore, proteins that play a role in mitosis are predicted to be potential targets for PGV-1 and piperine inhibitory effect and contribute to the incidence of mitotic catastrophe and senescence. The cytotoxic activity of 4T1 breast cancer cells and their effect on inducing mitotic catastrophe and senescence are investigated in this study. In addition, this study explores the target proteins of PGV-1 and piperine in TNBC and the molecular docking of these compounds against target proteins to reveal their inhibitory activity in TNBC.
2. Methods
2.1. Compounds
Piperine was acquired from Sigma-Aldrich (USA), while PGV-1 was synthesized by Cancer Chemoprevention Research Center, Faculty of Pharmacy, Universitas Gadjah Mada, Indonesia. Both compounds were dissolved in DMSO as a stock. Furthermore, the solutions were prepared in several concentrations in culture media.
2.2. Cell Culture
4T1 breast cancer cells (ATCC® CRL-2359) were provided by Professor Masashi Kawaichi, Nara Institute of Science and Technology, Japan. The cells were cultured in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, USA), HEPES (Sigma), 10% fetal bovine serum (FBS) (Gibco, USA) and 1% penicillin-streptomycin (Gibco, USA) under standard conditions (37°C, 5% CO2).
2.3. MTT Assay
A total of 7 × 103 4T1 cells/well were cultured into the 96-well plate and grown until confluent 80%. The cells were then treated with various doses of piperine (10 - 1000 µM) or PGV-1 (0.5 - 16 µM) or both at the selected concentrations and incubated for 24 h. After formazan crystals formed, the cells were rinsed with phosphate buffer saline (PBS) and then added with MTT reagent (Sigma) into each well and incubated for 4 h. Termination of the MTT reaction was carried out by adding a stopper reagent (0.01 N HCl containing 10% SDS), and then the plate was incubated overnight in dark conditions at room temperature. Furthermore, the absorbance of the plate was read by an ELISA reader (BioRad) at 595 nm. The absorbance data were then converted into percent cell viability to calculate the IC50 value (2). For the combined MTT assay, the IC50 values of the single treatment were referred to determine the concentration to be applied. There are three concentration series for each compound, i.e., 1/2 IC50, 1/4 IC50, and 1/8 IC50 (2). Cell viability data from the combined MTT assay were used to determine the Combination Index (CI) which is calculated based on Chou and Talalay's formula (15). The classification of CI values is CI < 1, = 1, and > 1 indicating synergism, additive effect and antagonism, respectively.
2.4. May Grunwald-Giemsa staining
4T1 cells with a density of 2 × 105 cells/well were grown into the six-well plate and incubated until confluent 80%. The cells were then treated with selected concentrations of piperine and PGV-1. After being incubated for 24 h, the cells were rinsed with PBS two times, and the May-Grünwald-Giemsa stain (0.25% w/v, in methanol) (Sigma-USA) was added to each well up to 5 min. After that, the cells were flushed with phosphate buffer pH 7.2 for 1.5 min, and a Giemsa stain diluted with phosphate buffer (1: 50) was added to the well. After waiting for about 15 to 20 min at room temperature, the cells were rinsed briefly by deionized water and finally air-dried. Observations were made using an inverted microscope (Leica) Mitotic catastrophe analysis is indicated by cells with micronucleus or polynuclear (3, 11). The percentage of mitotic catastrophe cells was calculated by two people with blinded observation method.
2.5. Senescence Associated-β-Galactosidase Assay
4T1 cells (2 × 105 cells/well) were seeded into the six-well plate then incubated until 80% confluent. The cells were treated by selected concentrations of piperine and PGV-1 for 24 h. After that, the cells were flushed by PBS twice, followed by the addition of a fixation buffer for 10 to 20 min, and then washed again with PBS once. Next, 1 - 2 mL of X-Gal solution is added to each well and incubated in a non-CO2 incubator at 37°C. The cells were observed after 72 h under an inverted microscope (Leica). The green color of the cells indicated senescence (3).
2.6. Analysis of Potential Target Proteins
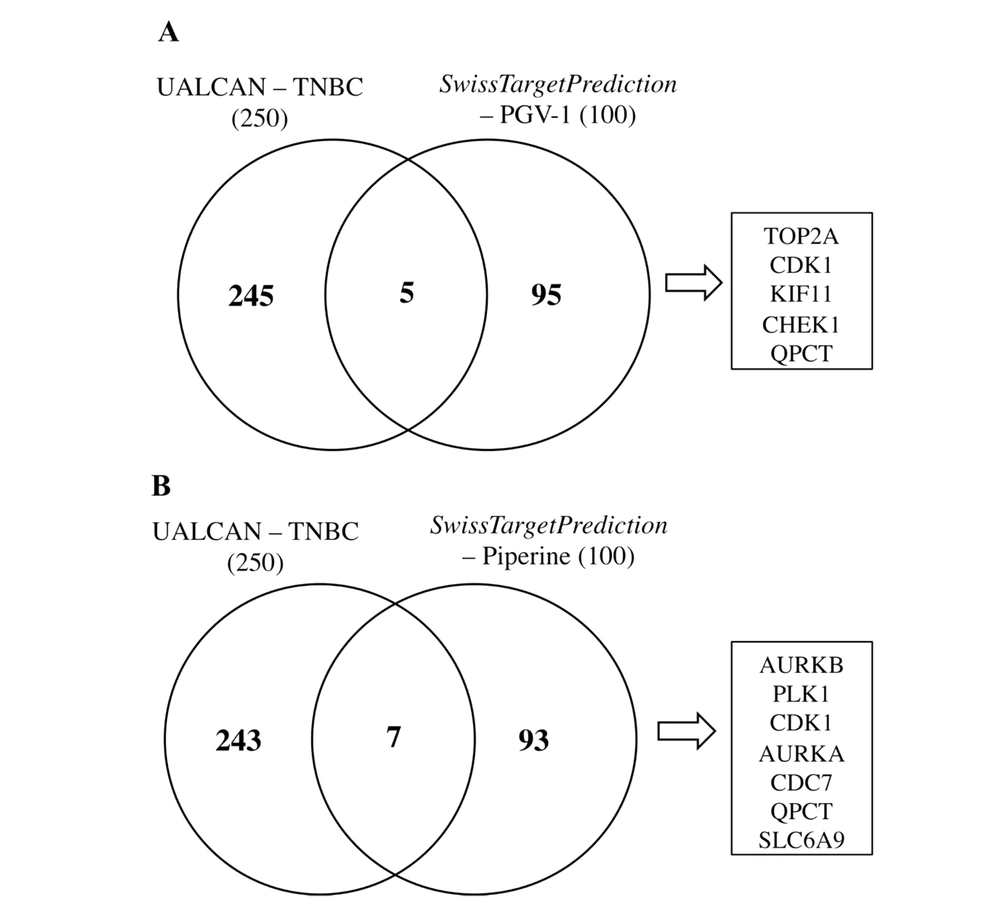
The predictive target protein was performed using SwissTargetPredicition (http://swisstargetprediction.ch/) by entering the SMILES code or chemical structure of PGV-1 or piperine. After that, a list of 100 target proteins is obtained (16). Subsequently, the list of overexpressed genes on TNBC is provided by the UALCAN database (http://ualcan.path.uab.edu/). A list of the highest 250 overexpressed genes is produced (17). Next, slices of the Venn diagram from 100 target proteins and 250 overexpressed genes are made to obtain the potential target PGV-1 or piperine in TNBC.
2.7. Expression of Target Genes
The target gene expression data on TNBC was carried out using the UALCAN database with TCGA analysis. The first step is to input the target gene in the query genes column then select "breast invasive carcinoma (BRCA)". After clicking "explore", the expression option on the page is explicitly selected on expression based on Major subclasses. Next, the data with statistical significance value and the visualization image were obtained (17).
2.8. The Survival Rate for Target Genes in Breast Cancer Patients
To determine the probability of survival, the OncoLnc database is used (http://oncolnc.org/). The first step is to submit the target gene and then select the type of cancer dataset, namely "BRCA". Next, we input the percentile column with 25. After that, the page showed the Kaplan plot images (18) and that data in an excel form.
2.9. Molecular Docking
A molecular docking investigation was used to confirm the binding relationship between PGV-1 and piperine toward CDK1, KIF11, AURKA, AURKB, and PLK1. To simulate molecule binding, calculate RMSD, and display protein-ligand interaction, a computational study was presented utilizing licensed software MOE 2010.10. CDK1, KIF11, AURKA, AURKB, and PLK1 had PDB IDs of 6GU6, 3ZCW, 2DWB, 2VGO, and 2YAC, respectively. The default settings were used, with the placement setting and scoring system being triangle matcher and London dG. The docking results from ten retain settings were refined using the force field approach (19). In ChemDraw software, the chemical structures of PGV-1 and piperine were produced, and then the structural energy was minimized, and a conformational structure was built in MOE. The molecular docking investigation was conducted with each protein's native ligand binding location in mind. The docking score and binding visualization of each drug with the target proteins were detailed in the molecular docking results.
2.10. Statistical Analysis
All data from triplicate measurements were evaluated using SPSS v.22 by one-way ANOVA accompanied by a Tukey HSD post-hoc test. The results are presented as the mean standard deviation of three independent experiments. P < 0.05 was used to determine statistically significant differences.
3. Results
3.1. The Anti-Proliferative Effect of Piperine and PGV-1 on 4T1 Cells
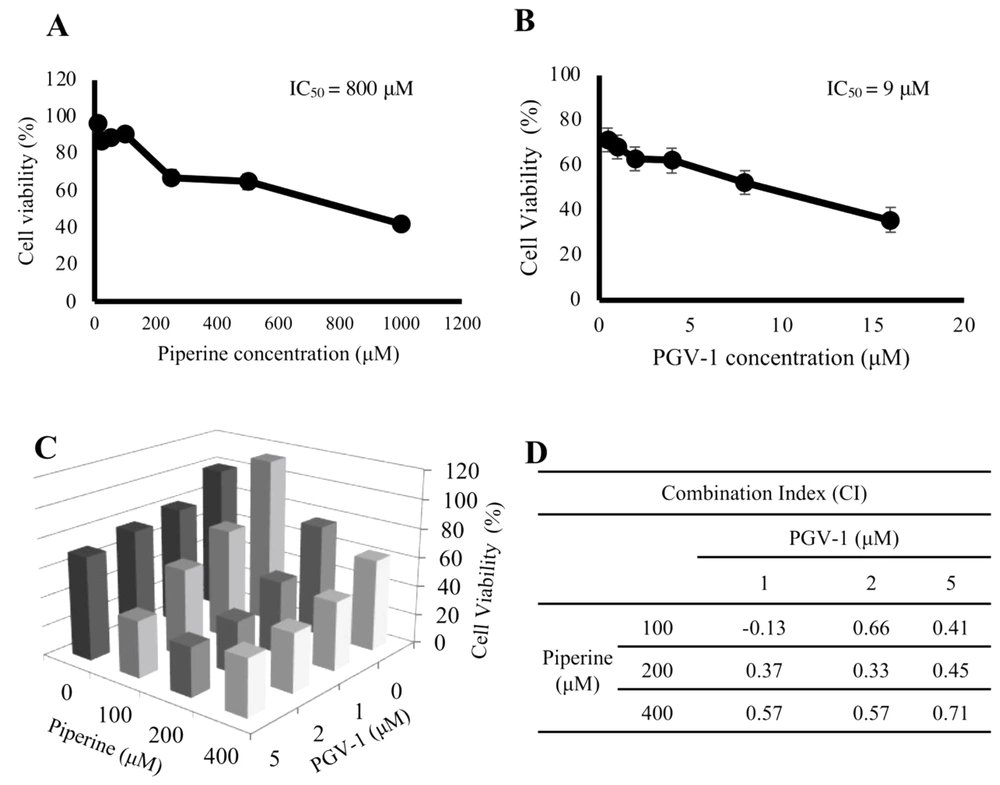
The piperine single cytotoxic assay on 4T1 cells showed an IC50 value of 800 µM (Figure 1A). Piperine cytotoxicity is not potent because the IC50 is above the 10-100 µM range (20). Meanwhile, the PGV-1 single cytotoxic assay results on 4T1 cells exhibited an IC50 value of 9 µM (Figure 1B). Therefore, the less potent cytotoxicity of piperine suggests that piperine has the potential to be promoted as a co-treatment (adjunctive) agent for PGV-1.
Cytotoxic effect of PGV-1 and piperine on 4T1 cells. 4T1 cells (7 × 103 cells/well) were cultured in a 96 well-plate and treated with PGV-1 and piperine for 24 h. A and B, The cytotoxicity of piperine and PGV-1 was expressed as percent cell viability (mean ± SD of 3 trials); C, The combination treatment showed lowering cell viability compared to a single treatment of each compound with IC50 of piperine (100, 200, 400 µM) combined with PGV-1 (1, 2, and 5 µM) for 24 h; D, Combination Index of PGV-1 and piperine.
The cytotoxic assay of the combination treatment, calculated by the CI, was used to determine the cytotoxic effect of the two compounds. CI values of < 1 indicate a synergistic effect, = 1 meaning additive and > 1 antagonistic (15). The concentrations of piperine and PGV-1 were used by using a combination concentration of IC50. The piperine concentrations included 100, 200, and 400 µM, while the concentrations of the PGV-1 were 1, 2, and 5 µM (Figure 1C). The results show that the combinations of PGV-1 with piperine showed a CI value < 1 (Figure 1D). These low CI values mean synergistic effects between piperine and PGV-1. Thus, piperine enhances the cytotoxic effect of PGV-1 on TNBC 4T1 cells.
3.2. The Effect of Mitotic Catastrophe Induction of Piperine and PGV-1 on 4T1 Cells
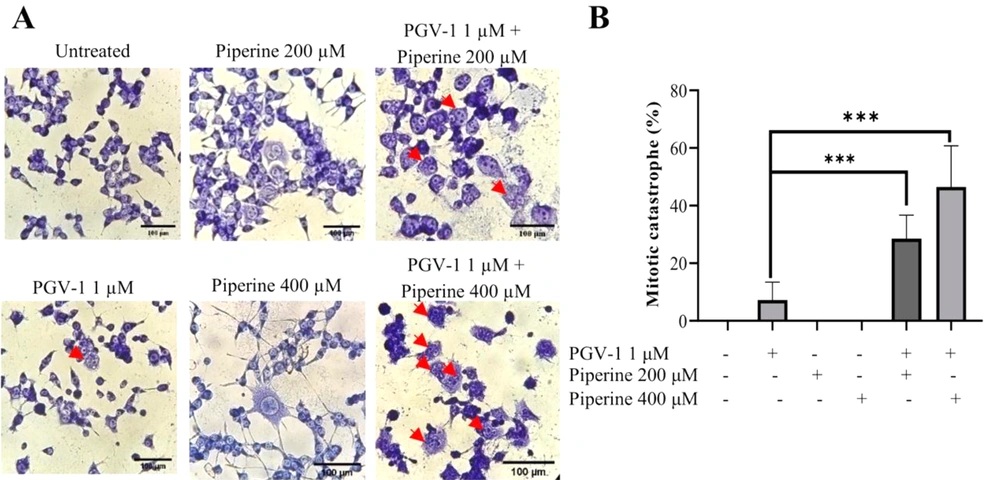
In some cell lines, PGV-1 induces cell cycle arrest at G2/M. We then observed whether the co-treatment of piperine and PGV-1 affect the mitotic evidence by using the May Grünwald-Giemsa staining. Two concentrations of compounds were used in this assay, namely 200 and 400 µM piperine and 1 µM PGV-1. The result showed that 1 µM PGV-1 induced polynuclear formation, indicating mitotic catastrophe occurrence shown by a red arrow (Figure 2A). Interestingly, the number of polynuclear cells was found to be significantly higher (P < 0.001) when PGV-1 1 µM was combined with piperine 200 and 400 µM at 29 and 46%, respectively (Figure 2B). These results indicate that the triggering of mitotic catastrophe is more effective with combination treatment than a single treatment.
Mitotic catastrophe induction effect of PGV-1 and piperine on 4T1 cells. 4T1 cells (2 × 105 cells/ well) were grown in a 6 well-plate and treated with PGV-1 and piperine for 24 hours. Mitotic catastrophe cells were analyzed using the May Grünwald-Giemsa staining assay under an inverted microscope. A, The morphology of cells of the several treatments is indicated. The red arrows indicate the polynucleated cells/mitotic catastrophe. B, The percentage of mitotic catastrophe cells from microscopic observation. Data are expressed as mean ± SD of three independent experiments (*** P < 0.001).
3.3. The Effect of Senescence Induction of Piperine and PGV-1 on 4T1 Cells
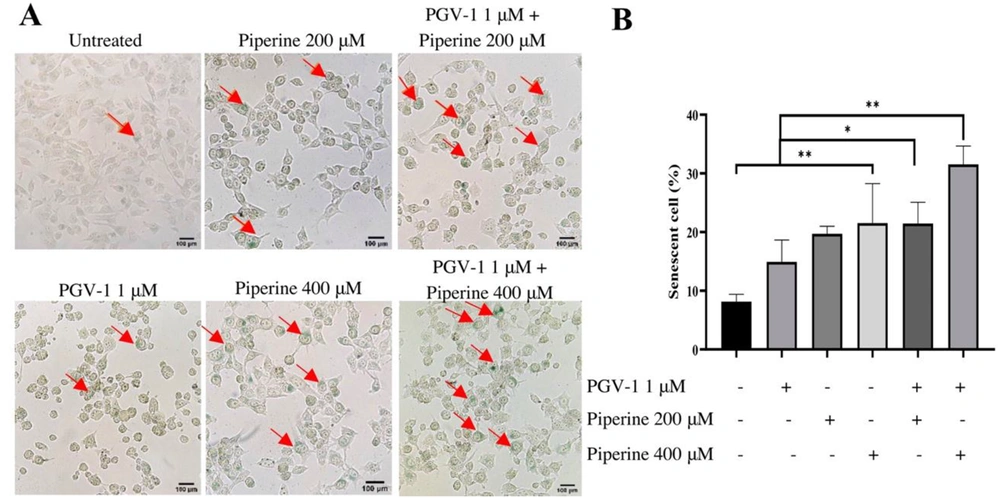
Mitotic catastrophe is a type of cell cycle arrest occurring during the mitotic phase closely related to senescence evidence. We then performed an SA-β-Galactosidase assay with a single treatment and combination of PGV-1 and piperine to determine the effect in inducing senescence. The senescence assay of 4T1 cells with a single treatment showed that the concentration of 1/2 IC50 µM piperine induced higher senescence than control cells and 1/4 IC50 piperine. Likewise, the 1 µM PGV-1 induced higher senescence than control cells but lower than the 200 and 400 µM of piperine (P < 0.05) (Figure 3B). The number of senescent cells in the 1 µM PGV-1 combination treatment with 200 µM piperine increased by 8% compared to PGV-1 1 µM alone and increased by 17% (P < 0.05) to control cells. Furthermore, the combination treatment of PGV-1 1 µM with piperine 400 µM gave a higher senescent cells effect. In this combination treatment, the number of senescent cells increased by 10% from the combination treatment with 200 µM piperine (P < 0.05) (Figure 3B). Thus, the combination treatment of PGV-1 with piperine was able to increase senescence induction in 4T1 cells.
Induction effect of PGV-1 and piperine senescence cells on 4T1 cells. 4T1 cells (2 × 105 cells/ well) were seeded in a 6-well plate and treated with PGV-1 and piperine for 24 hours then observed after 72 hours. Senescent cells were analyzed using the SA-β-galactosidase staining assay under an inverted microscope. A, Morphological appearance of the cells in the several treatments as indicated. The red arrows indicate the senescent cells. B, The percentage of senescent cells from microscopic observation. Data are expressed as mean ± SD from triplicate independent trials (* P < 0.05, ** P < 0.01).
3.4. Prediction of Piperine and PGV-1 Target Proteins
All those results indicate that PGV-1 and piperine synergistically induce cell aging, which leads to apoptosis and cell death. However, the explanation regarding the molecular mechanism related to the essential proteins that target these two compounds has not been studied further. We then investigated the target proteins underlying the synergistic co-treatment of both compounds by employing bioinformatics analysis.
The piperine and PGV-1 predictive target genes were searched under the SMILES code using the SwissTargetPrediction online database, which provides an accurate set of predictive target gene results. Then all predictive target genes/proteins were aligned with the TNBC breast cancer overexpressed gene list obtained from the UALCAN database to match these genes/proteins. The proteins in the sliced diagram were taken as prospective target proteins for piperine and PGV-1 for TNBC breast cancer therapy.
Venn diagrams were used to determine the total sliced target proteins of the predictive target genes for piperine and PGV-1 with a list of overexpressed genes in TNBC. We found that 250 genes are overexpressed in breast cancer based on the TNBC category. Meanwhile, the SwissTargetPrediction search results found 100 PGV-1 and piperine predictive target genes each. The results of the Venn diagram slice yielded five potential target genes from UALCAN gene alignment with PGV-1 target gene prediction, namely TOP2A, CDK1, KIF11, CHEK1, and QPCT (Figure 4A) and seven potential target genes with piperine target gene prediction, namely AURKB, PLK1, CDK1, AURKA, CDC7, QPCT, and SLC6A9 (Figure 4B). Each of these genes has a different function in regulating cell work, but interestingly, most of these genes play a role in the cell cycle (Table 1). The cell cycle is an appropriate target in cancer treatment and matches the finding in this study. Therefore, the following discussion focuses on several target proteins that take a role in mitotic regulation.
Predictive target proteins of PGV-1 and piperine in TNBC. 250 proteins overexpressed in TNBC were obtained from UALCAN, while 100 target proteins of PGV-1 and piperine were respectively obtained from SwissTargetPrediction. A, Protein slice between the overexpressed protein in TNBC and the predictive target protein PGV-1; B, Protein slice between the overexpressed protein in TNBC and the predictive target protein piperine. Venn diagram analysis using InteractiVenn.
| No. | Protein Name | Role | Function | Reference |
|---|---|---|---|---|
| 1 | AURKB | Protein kinase: phosphorylates CHMP4C and complex subunit CPC | Regulation of chromosome alignment during mitosis by association with microtubules | (21, 22) |
| 2 | PLK1 | Protein kinase phosphorylates CEP170, CDC25, CCNB1, and KIF20A | Controls centrosome maturation and spindle formation., inactivation of complex/ cyclosome inhibitors (APC/C) | (23, 24) |
| 3 | CDK1 | Forming complex proteins with Cyclin B: phosphorylates CEP1669, CDC7, CDC25, and EML3 | Initiate mitosis, the G2-M transition, and organize G1 and G1-S by assemblage with multiple interphase cycles. | (25, 26) |
| 4 | AURKA | Protein kinase: phosphorylates CDC25B and KIF2A | Interacts to the centrosome and spindle microtubules during M-phase | (27, 28) |
| 5 | CDC7 | Protein kinase: phosphorylates MCM 2 and MCM 3 | Regulation of G1-S transitions and chromosome replication | (29) |
| 6 | QPCT | Synthesis of pyro-glutamyl peptides. | Regulates pathways related to the innate immune system | (30) |
| 7 | SLC6A9 | Transport protein | Regulates glycine levels in neurotransmission mediated by NMDA receptors. | (31) |
| 8 | TOP2A | Binds to two double-stranded DNA molecules | Chromosome condensation, chromatid separation, and regulation during DNA transcription and replication. | (32) |
| 9 | KIF11 | Interacts with NEK6 | Chromosome placement, centrosome segregation, and formation of bipolar shafts during the M phase. | (33) |
| 10 | CHEK1 | Protein kinase: phosphorylates CDC25A | Checkpoints in reaction to DNA damage or the presence of non-replicated DNA. | (34) |
3.5. Expression of Potential Piperine and PGV-1 Target Protein in Breast Cancer
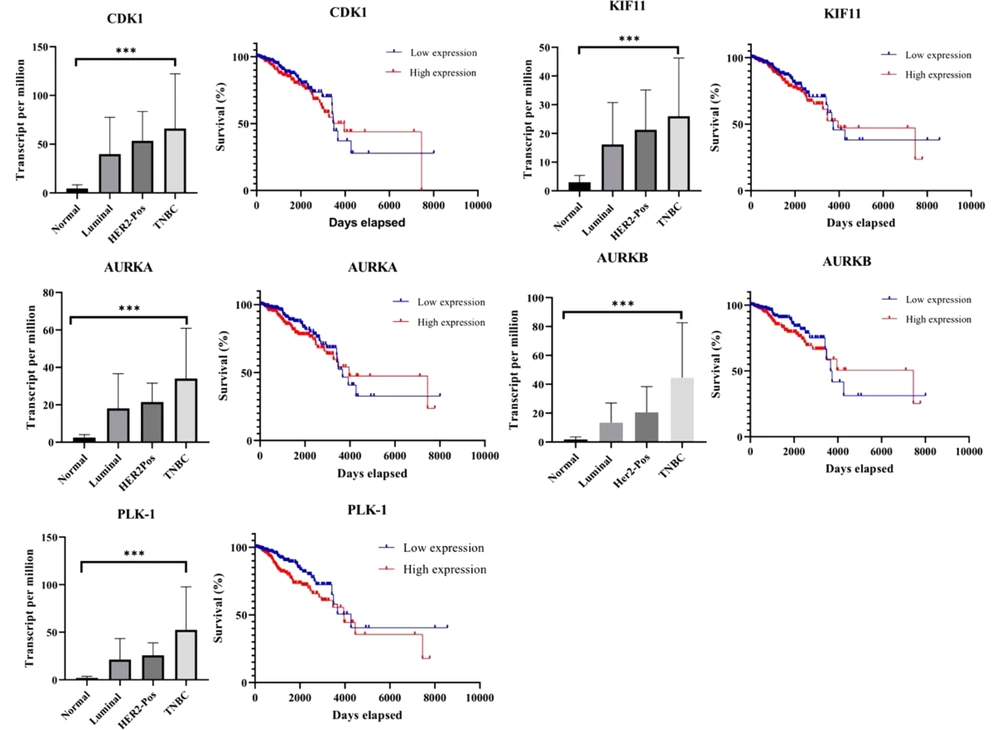
We noted that most of the target proteins of PGV-1 and piperine play a role in the mitotic progression, namely CDK1, KIF11, AURKA, AURKB, and PLK1. We explored the gene expression profiles in BRCA (breast invasive carcinoma) breast cancer from the TCGA database via UALCAN. The retrieved data from TCGA showed that all potential genes/proteins were overexpressed in tumor tissue and significantly (P < 0.01) expressed higher than normal tissue indicated by the transcript value. Further analysis showed that breast cancer with TNBC subtypes showed the highest transcript value compared to other subtypes such as luminal and Her2 positive in almost all potential target genes (Figure 5). This fact supports our finding that PGV-1 and piperine may share a target to induce mitotic arrest on 4T1 cells.
Expression of PGV-1 and piperine target proteins on BRCA and their correlation with Survival of BRCA patients. Data of target gene expression based on the breast cancer subclass were obtained from UALCAN (number of samples: normal = 114, luminal = 566, HER2-pos = 87, and TNBC = 116), while the data of the effect of target gene expression level on BRCA patient survival were obtained from OncoLnc (number of samples: low expression = 251 and high expression = 251) (*** P < 0.001).
3.6. Effect of Target Protein Expression on Breast Cancer Patient Survival
Overexpression of the target protein for piperine and PGV-1 in the TNBC subtype breast cancer correlates with the survival of breast cancer patients. We performed an overall survival analysis via the OncoLnc website. Piperine and PGV-1 targeted protein products (CDK1, KIF11, AURKA, AURKB, and PLK1) were found to have a significant (P < 0.05) impact on overall survival duration (Figure 5). This suggests that patients with higher CDK1, KIF11, AURKA, AURKB, and PLK1 protein expression in their tissues have a considerably shorter overall survival than those with lower expression.
3.7. Screening PGV-1 and Piperine Target Protein by Molecular Docking
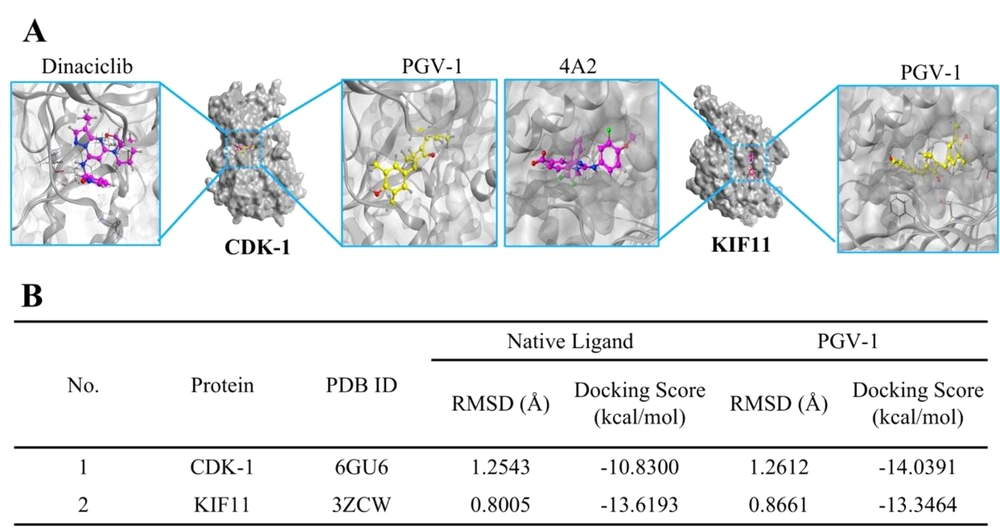
Our screening revealed that binding of PGV-1 to CDK1 and KIF11 exhibited lower docking scores (-14.0391 and 13.3464 kcal/mol, respectively) than native ligands Dinaciclib and 4A2, respectively (Figure 6B). Furthermore, piperine binds to the binding site of the target protein and interferes with protein-protein interactions (Figure 6A). Taken together, the results reveal that CDK1 and KIF11 are promising targets for PGV-1.
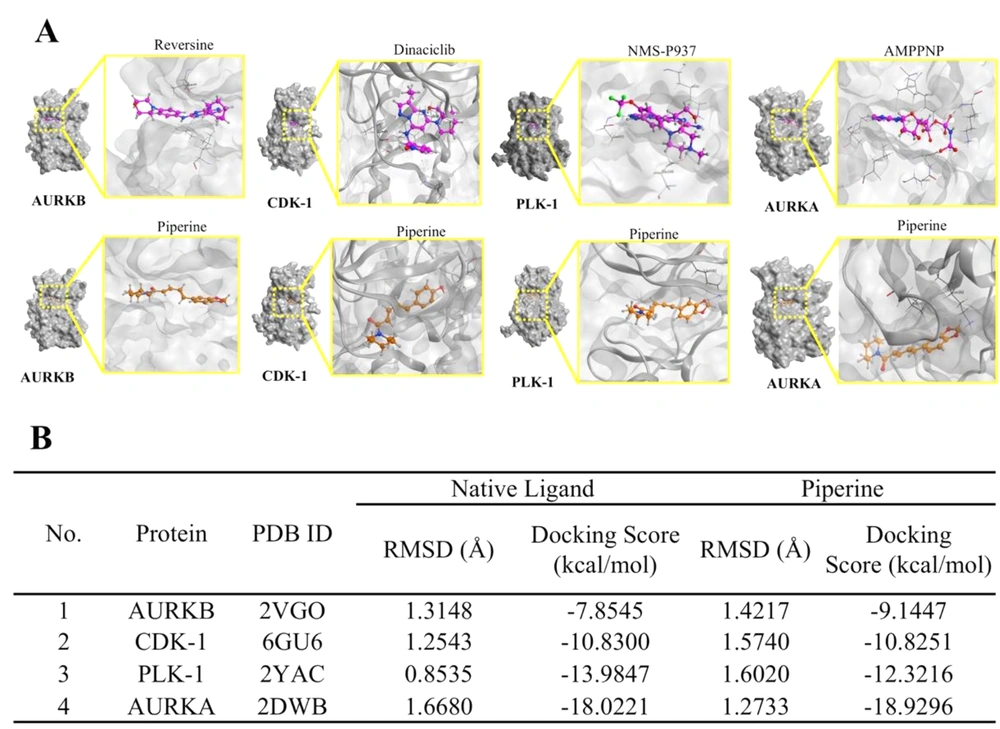
As shown in Figure 7B, our screening results revealed that the bonding of piperine with AURKA showed the lowest docking score (-18.9296 kcal mol), which means that AURKA had the highest binding piperine. Furthermore, PGV-1 binds AURKA slightly better than Reversine (a native ligand) with a docking score of -18.0221. Meanwhile, piperine binds to AURKB with the highest docking score of -9.1447 kcal/mol. However, this score is relatively lower when compared to the bond between AURKB and Dinacicilib (-7.8545 kcal/mol). Other target proteins such as CDK1 and PLK1 showed lower affinity than AURKA, but higher than AURKB.
Meanwhile, the affinity of piperine for these two proteins did not show a significant difference compared to their respective native ligands. Furthermore, piperine binds to the binding site of the target protein and interferes with protein-protein interactions (Figure 7A). Taken together, the results reveal that AURKA, AURKB, CDK1, and PLK1 are the most promising protein targets and essential structures to consider for further strain screening.
4. Discussion
The curcumin analog PGV-1 has been known to have a cytotoxic effect on several cancer cell cultures (1). Exploration of PGV-1 as a potential anti-cancer candidate provides new hope for cancer treatment because of its low side effects on normal cells (2, 3). Thus, the effectiveness of PGV-1 needs to be optimized through co-treatment with natural-based compounds such as piperine because of its ability to increase the effect of a therapeutic agent (35). The results of this study exhibited that the single treatment of PGV-1 exhibited an IC50 value of 9 µM and piperine 800 µM in 4T1 cells. The IC50 value of the PGV-1 is higher than the previous research, namely 4 µM at 24-hour treatment (2). Likewise, piperine in previous studies had a lower IC50 value, which was 105 µM at 48 h of treatment (8). This difference in IC50 value is possible because of the different situations and conditions of the research. However, in essence, PGV-1 has a more potent cytotoxic effect than piperine, resulting in a synergistic cytotoxic effect when combined.
The combination assay showed that the co-treatment of PGV-1 with piperine had more toxic activity against 4T1 cancer cells than the single treatment. Piperine was able to increase the effectiveness of PGV-1 by showing a synergistic effect with a CI value < 1. In line with previous studies, piperine escalates the anti-cancer activity of curcumin toward HCT116 cells (6). Thus, piperine is prospective to be applied as an adjunctive therapy agent for PGV-1.
The results of the cytotoxic assay were then continued by exploring the induction of cell cycle inhibition. PGV-1 can inhibit the cell cycle in the G2/M phase (2) and are more specific in pro-metaphase (3). In line with the results of this study, based on the May Grünwald-Giemsa staining, it was found that PGV-1 and its combination with piperine inhibited the cell cycle in the mitotic phase through mitotic catastrophe events in 4T1 cells. Mitotic catastrophe is represented by typical nuclear changes and the formation of polynuclear giant cells (36). These polynuclear giant cells then undergo gradual cell death through necrosis or apoptosis (37). These prominent markers appeared in 4T1 cells after being treated with 1 µM PGV-1. However, the highest incidence of mitotic catastrophe was in the combination of PGV-1 1 µM with 200 µM and 400 µM of piperine. Since most cells that undergo mitotic catastrophe eventually die, the cellular process leading to a permanent cessation of growth is referred to as a condition known as senescence (13).
Both PGV-1 and piperine, with alone treatment or in combination, can induce senescence on 4T1 cells. However, the combination of PGV-1 1 µM with piperine 200 µM and 400 µM increased senescence up to 8 to 18% compared to 1 µM PGV-1 treatment alone. The mechanism of aging induction is predicted by activating the p16/INKa and/or pRB/CDKN2A pathways (38). P16/INKa integrates signals for DNA damage-induced senescence that imposes CDK1-mediated cell cycle inhibition (39). It is known that PGV-1 accumulates cells in the pro-metaphase phase and induces senescence, similar to one of the functions of p16/INKa (3).
DNA damage caused by genotoxic agents activates Chk1, which prevents mitotic entry through inhibition of CDK1 activity, affecting the decrease in cyclin B1 (10), resulting in temporary accumulation of cells in the G2 phase, but ultimately dies during entry into mitosis (11). This phenomenon is known as mitotic catastrophe (12). The mitotic catastrophe event characterized by micronuclei formation further supports this evidence that the valid target of PGV-1 is not G2 inhibition but rather mitosis. This is in line with the previous revealing that PGV-1 decreased Cyclin B1 expression (2). The results of this study indicated that treatment with PGV-1 suppressed the mitotic cycle in 4T1 cells. Furthermore, how the mechanism of PGV-1 and piperine causes mitotic catastrophe molecularly and the potential target proteins that are affected by PGV-1 and piperine are further explored through bioinformatics studies.
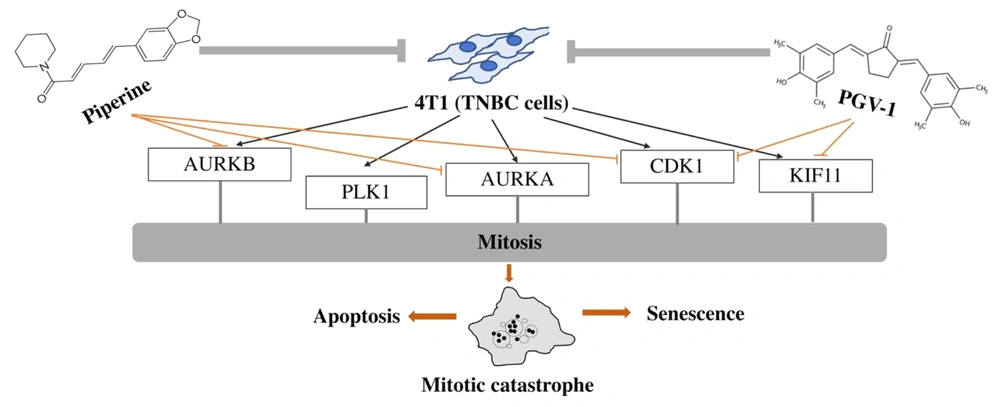
The analysis results under UALCAN and SwissTargetPrediction found as many as five proteins as targets of PGV-1 and seven proteins as targets of piperine. Interestingly, PGV-1 and piperine target different proteins, which further provides evidence that these two compounds can cooperate synergistically. PGV-1 target proteins include TOP2A, CDK1, KIF11, CHEK1, and QPCT. Meanwhile, AURKB, PLK1, CDK1, AURKA, CDC7, QPCT, and SLC6A9 are potential target proteins for piperine. Further analysis showed that these proteins largely contribute a role in the cell cycle. In line with the initial study finding that PGV-1 triggers cell cycle arrest in the G2/M phase (2) and, more specifically, in pre-metaphase (3). It is also in line with piperine which causes cell cycle termination at more varied phases, namely G0/G1 in 4T1 cells and G2/M in HeLa cells (8, 40).
Of these target proteins, five main proteins that regulate the cell cycle, especially in mitosis, namely CDK1, KIF11, AURKA, AURKB, and PLK1, were analyzed further. The gene expression analysis of the target protein in BRCA cells showed that the target protein was expressed higher than normal cells. Furthermore, based on breast cancer subtypes, it is known that the target protein is expressed higher in TNBC subtypes than other breast cancer subtypes such as luminal and HER2-positive subtypes. Overexpression of these target genes also correlated with survival rates of BRCA patients. In harmony with Jiang et al. (2017) revealed that KIF11 is overexpressed in TNBC, and its expression is correlated with shorter survival times, and so is AURKB (41). Therefore, therapeutic targeting of some of these proteins is expected to suppress expression and increase survival rates.
In the cell cycle, CDK1 and Cyclin B combine to form a protein complex that is very important in initiating G2/M (25). Targeting CDK1 as a TNBC marker is one anti-cancer mechanism through synthetically lethal which refers to mutations in one of two genes that are compatible with viability but mutations in both cause death (14). Furthermore, KIF11 plays a role in positioning chromosomes, separating centrosomes, and forming a bipolar axis during mitosis (33). Inactivation of KIF11 causes defective cell division and cell cycle arrest during mitosis, resulting in apoptotic cell death (41). AURKA and AURKB (aurora kinase A and B) play a role in regulating chromosomal alignment during mitosis through association with centrosomes and spindle microtubules during mitosis (22, 28). AURKA and AURKB are overexpressed in TNBC, associated with polymorphism, and are associated with survival rates (42). Furthermore, PLK1 plays a role in regulating centrosome maturation and spindle assembly and inactivation of inhibitory complexes/cyclosomes (APC/C) (24). Therefore, targeting PGV-1 and piperine on CDK1, KIF11, AURKA, AURKB, and PLK1 demonstrated the potential for PGV-1 and piperine to synergize in the cell cycle arrest characterized by deregulation of mitosis or mitotic catastrophe.
The results of the molecular docking screening in this study found that CDK1 is a more desirable candidate protein than KIF11 to bind to PGV-1. PGV-1 shows a binding affinity with CDK1 that is superior to the inhibitor dinaciclib. Also, piperine can be predicted to be an inhibitor of AURKB, which has a stronger affinity compared to reversine. Overall, PGV-1 and piperine have potential as inhibitors of CDK1, KIF11, AURKA, AURKB, and PLK1 proteins.
Overall, this study revealed the synergistic effect of the combination of piperine with PGV-1 on 4T1 TNBC cells. The mechanism of the synergistic effect of PGV-1 and piperine on 4T1 TNBC cells was further demonstrated that both target different proteins. Of these target proteins, five proteins have a pivotal role in the cell cycle, namely CDK1 and KIF11, specific targets for PGV-1 and AURKA, AURKB, and PLK1, which are specific targets for piperine. The synergistic effect of PGV-1 was further proven through the mitotic catastrophe event, which was also predicted to be caused by inhibition of mentioned proteins. Piperine strongly exhibits a potential to be applied as adjunctive TNBC cancer therapy with PGV-1.
4.1. Conclusion
Overall, piperine demonstrated a synergistic effect with PGV-1 on 4T1 cells related to mitotic catastrophe and senescence. In addition, some proteins that may be targets of PGV-1 and piperine in correlation with are CDK1, KIF11, AURKA, AURKB, and PLK1.