Introduction
Cyanobacteria (blue-green algae) are a group of prokaryotes with photosynthetic properties. Unicellular and filamentous structures are common forms of cyanobacteria (1). They are valuable organisms in aquatic and terrestrial ecosystems. In addition, cyano-bacteria can fix atmospheric nitrogen by converting it into ammonium compounds (2). Cyanobacteria can survive in different habitats and produce various secondary metabolites with valuable and distinctive functions. Among them, photoprotective metabolites such as scytonemin and mycosporine-like amino acids (MAAs) play a remarkable role in their protection against ultraviolet radiation (3). MAAs are colorless and water-soluble small molecules with a cyclohexenone or cyclohexenimine chromophore linked through sulfate esters or glycosides bond to imine functional group of amino acids or amino alcohols (4, 5). Based on the evidence, their biosynthesis takes place through the shikimate and/or pentose phosphate pathways (6). They can absorb light in the range of UV-A and UV-B wavelength (310-362 nm) (7). Due to UV absorption in a broad range of wavelength, high molar extinction coefficient (ε = 28,100–50,000 M-1cm-1), strong UV-absorption ability, and photostability, MAAs are applied as sunscreen agents in pharmaceutical and cosmetic products and can inhibit skin damage after UV irradiation (5, 8), such as Helioguard 365™ that contains Shinorine and Porphyra-334 (two well-known MAAs) as sun-protective agents (9). MAAs can also have antioxidant properties to scavenge toxic oxygen radicals and protect cells against oxidative stress (10, 11). Several studies have been shown that MAAs could protect cells against DNA damage and could block thymine dimer formation induced by UV irradiation. Moreover, it was found that MAAs promoted the proliferation of human skin fibroblast cells (12). This feature makes MAAs an attractive ingredient in cosmetics.
The presence of MAAs in a number of cyanobacteria species has been studied so far. In addition, the induction of MAAs in different types of cyanobacteria has also been investigated (13, 14). For example, the cyanobacterium Fischerella muscicola TISTR8215 was shown to produce UV-absorbing MAAs with maximum absorption at 332 nm and a retention time (Rt) of around 16.1 min, which was induced by UV radiation (13). Accordingly, two species of Fischerella sp. was investigated in this study regarding their MAAs production in normal condition (BG11 culture medium, without extra UV irradiation).
Experimental
Identification of cyanobacteria strains
Cyanobacteria samples (F5 and F14) had collected from the North of Iran. Morphological determination of samples was performed at the Department of Biotechnology, School of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran (15). For molecular confirmation of these two species of cyanobacteria, 16S rRNA ribosomal gene sequencing was applied.
Cyanobacteria cells (1 mL) were harvested from culture media by centrifugation (SV 1422, Sigma, Germany) at 10000 ×g for 1 min in sterile microtubes. Pellets were suspended in 100 µL of 10% Triton X100, vortexed, and incubated in a dry block incubator (DPI-1T, Kiagen, Iran) at 100 ºC for 60 min. The samples were cooled, vortexed (MS 3 B, IKA, Korea) and centrifuged (10000 ×g, 1 min, and 4 ºC); then, the supernatant of each sample was used as a DNA template. PCR amplification was performed in a total volume 50 µL containing 1 µL of DNA template in Master Mix 2x and 1 µL of general primes (Table 1). The PCR was run in a thermal cycler (T100™, BIORAD, USA) using the following protocol: initial denaturation at 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 57 °C for 30 s, 72 °C for 45 s, and an extension step at 72 °C for 20 min.
Amplified PCR products were analyzed using 2% agarose gel electrophoresis (Mini horizontal, Clever, UK) in comparison with the 1.5 kbp DNA ladder. Then, the amplified products were purified from agarose gel using the AccuPrep Gel DNA Purification kit (Bioneer, Korea). Finally, the sequences of amplified genes were determined using the standard Sanger method and analyzed using NCBI GenBank and BLAST program.
Cultivation and determination of growth curve of cyanobacteria
Both Fischerella sp. (F5 and F14) were cultivated in 500 mL Erlenmeyer flask containing 200 mL BG-11 medium and incubated at 25 ºC in germinator under a 16:8 h light/dark program and 70% humidity during 21 days. Two-milliliter culture samples (F5 and F14) were transferred to Falcone tubes containing 20 mL BG-11 and were incubated in the same condition. Then, the cultures were centrifuged at 10000 ×g for 10 min on alternate days, and the obtained biomasses were dried at 60 °C in an oven overnight and then weighed. Growth curves were drawn based on dry biomass weight and growth period using GraphPad Prism.
MAA extraction from Cyanobacteria
MAAs were extracted using methanol (MeOH) as described in previous studies (18, 19). Briefly, the cyanobacteria biomasses were separated from 200 mL culture of cyanobacteria by centrifugation at 10000 ×g for 10 min. Pellets were suspended in distilled water and re-centrifuged to remove impurities. These steps were repeated twice and then preserved in a freezer (-80 ºC) overnight and dried using a freeze-dryer (Alpha 1-2 LDplus, Christ, Germany). Then, 10 mL of absolute MeOH were added to 30 mg of dried biomasses. The mixtures were maintained in an ultrasonic bath (1 min) and then refrigerated. After 24 h, the samples were incubated in an ultrasonic bath at 45 ºC for 2 h and then centrifuged (10000 ×g, 15 min, and 4 ºC). Supernatants were collected and dried at room temperature and followed by dissolution in distilled water (1 mL). These samples were centrifuged again, and supernatants were transferred to the new microtubes. After adding 500 µL of chloroform to each sample, they were vortexed and centrifuged (10000 ×g, 10 min, and 4 ºC), and supernatants were collected for HPLC analysis.
HPLC analysis
Samples were analyzed by reverse-phase high-performance liquid chromatography (LC-8A, Shimadzu, Japan) equipped with a photodiode array (PDA) detector (SPD-M10A- Shimadzu). 100 µL of each were subjected to HPLC column (C18, 5 µm, 250 × 4.6 mm) through an isocratic mobile system of MeOH (25%) and 0.1% acetic acid with a flow rate of 1 mL/min for 20 min and column eluting compounds were detected in 340 nm.
LC-MS analysis
The LC-MS chromatograms of samples were recorded on HPLC (Agilent 1200 series) equipped with Agilent 6410 QqQ mass spectrophotometer with an electrospray ionization (ESI) interface. Samples (200 µL) were injected into the HPLC column (C18, 5 µm) using the same isocratic flow system with a flow rate of 1 mL/min for 10 min. In addition, the absorbance of eluting compounds was recorded at 320 nm.
Results
Identification of cyanobacteria genus
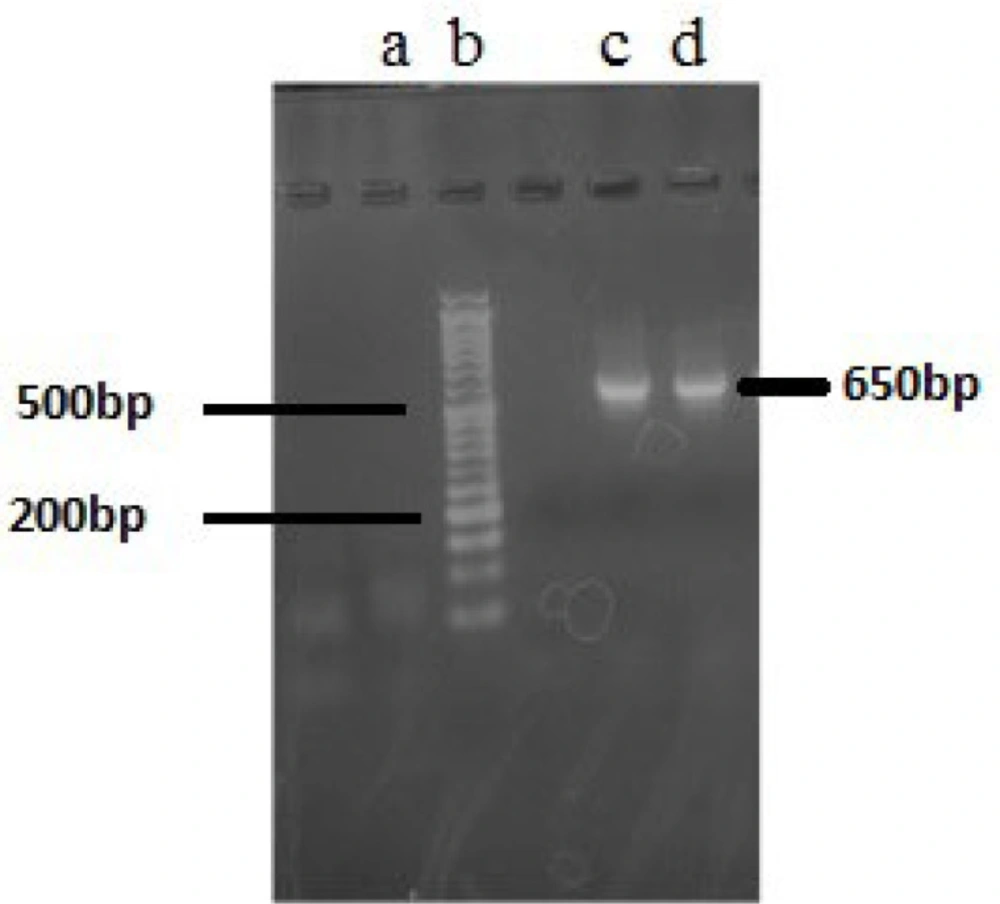
Based on the morphological assessment, two cyanobacteria strains (F5 and F14) studied in this research belonged to the Fischerella species. However, PCR sequencing of 16S rRNA genes as a molecular genetic approach was applied for more confirmation. As shown in Figure 1, PCR amplification with two specific primers of CYA106F/ CYAN738R, which results in 650 base pairs amplicon, showed acceptable results. Comparison of gene sequencing results with those available in the NCBI database showed that all of the obtained sequences corresponded to Fischerella sp. with high sequence similarity (99.6 %) (Table 2).
Growth measurements studies
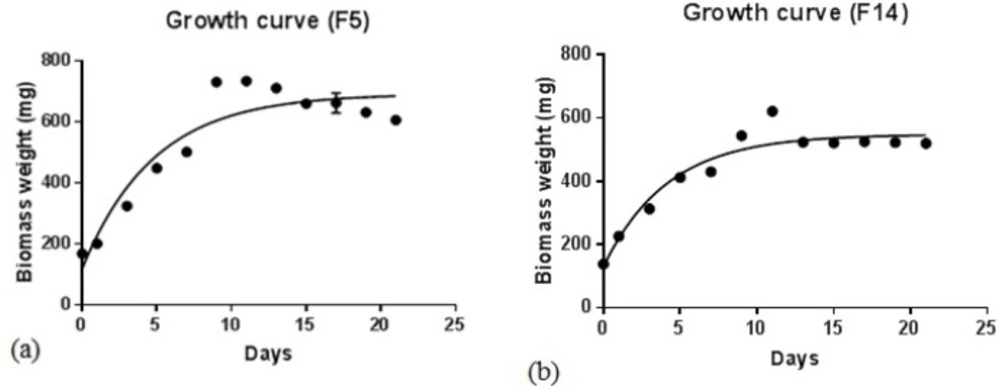
Fischerella strains (F5 and F14) were cultured in BG11 medium (200 ml), and then growth curves were drawn by the measurement of dried biomasses at 25-day intervals. As shown in Figures 2A and 2B, both species entered the logarithmic phase of growth after 15 days, and then the steady-state phase continued to 25 days. For more production of secondary metabolites, the extraction process was carried out after 21 days.
HPLC analysis of MAA partial extracts
Due to the high polarity of MAAs, they were extracted from F5 and F14 species biomass with 100% methanol. The presence of MAAs in these extracts was investigated by HPLC equipped with a PDA detector. In general, mycosporines with a common cyclohexenone ring system and a methoxy moiety at carbon 2 exhibited similar spectral characteristics, which had a maximum absorption (λmax) at about 310 nm in H2O. Different structures of MAAs which have identical ring-chromophores also had very similar spectral characteristics. Most marine MAAs are imine derivatives of mycosporines containing an amino-cyclohexenimine ring linked to an amino acid, amino alcohol, or amino group and have a maximum absorption between 320 and 360 nm (20).
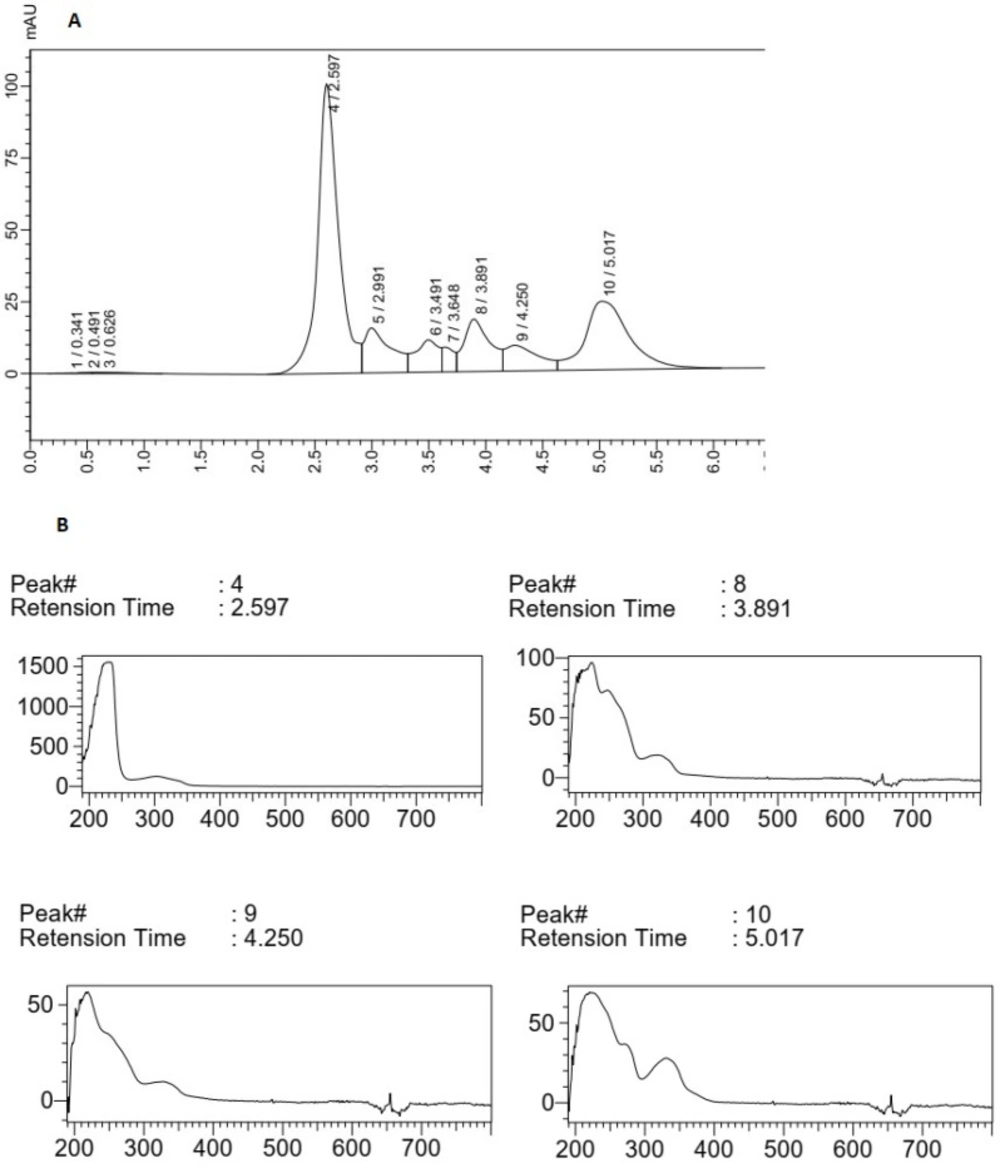
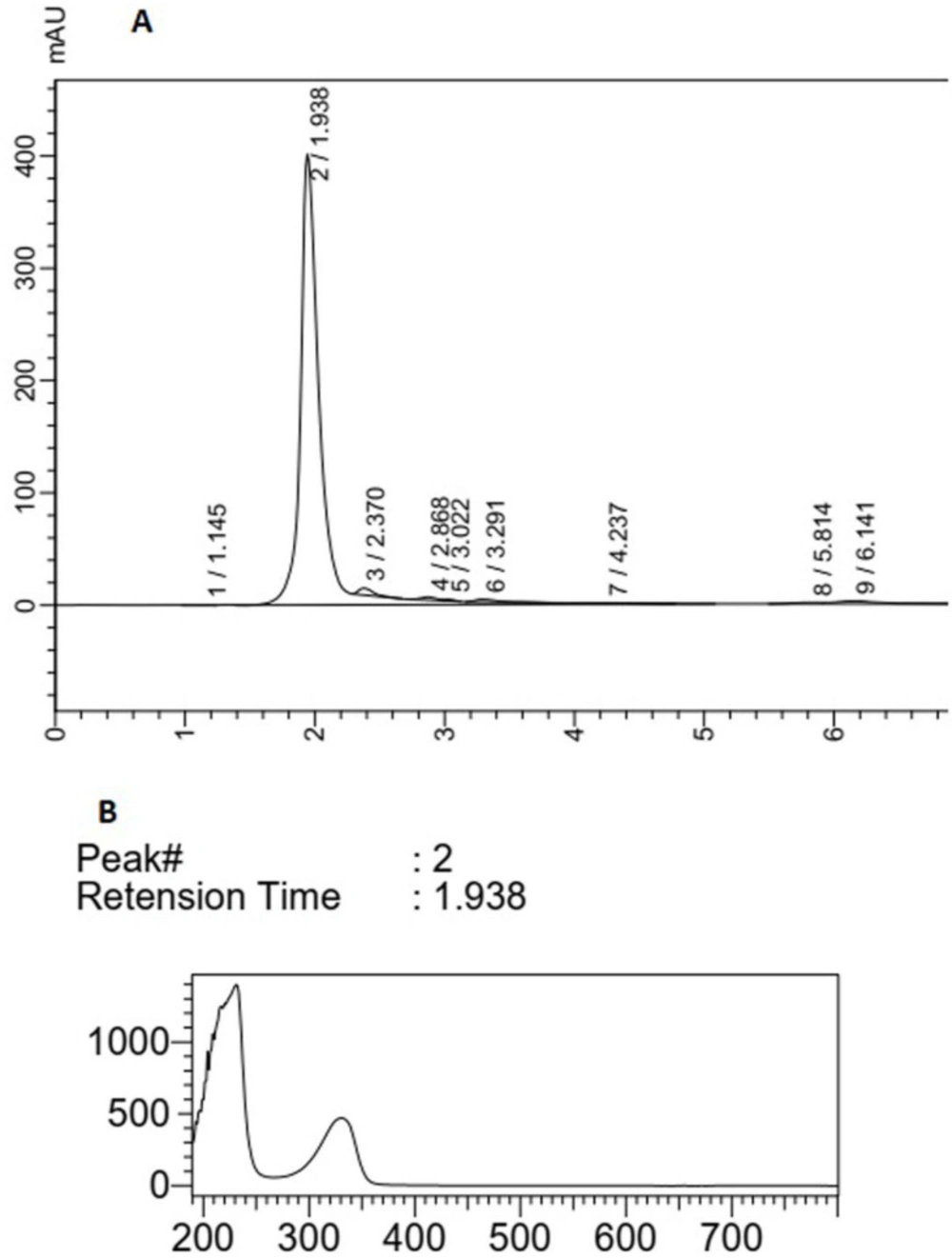
The HPLC chromatogram of F5 extract displayed a major peak (Rt of 2.597 min) with maximum absorption of about 270 nm and three peaks (Rt of peaks were 3.89, 4.25, and 5.01 min) with maximum absorption at 310-330 nm, which could show the MAA compounds (Figure 3). The main peak (Rt: 1.93 min) with λ max = 330 nm in the chromatogram of F14 extract could also confirm the production of MAAs in this species (Figure 4). The supernatant of Fischerella sp. cultivation mediums was similarly extracted by methanol (100%) and analyzed using HPLC. There was no evidence about any compound with maximum adsorption of 310-360 nm in these samples (data was not presented).
LC-MS analysis of cyanobacteria extracts
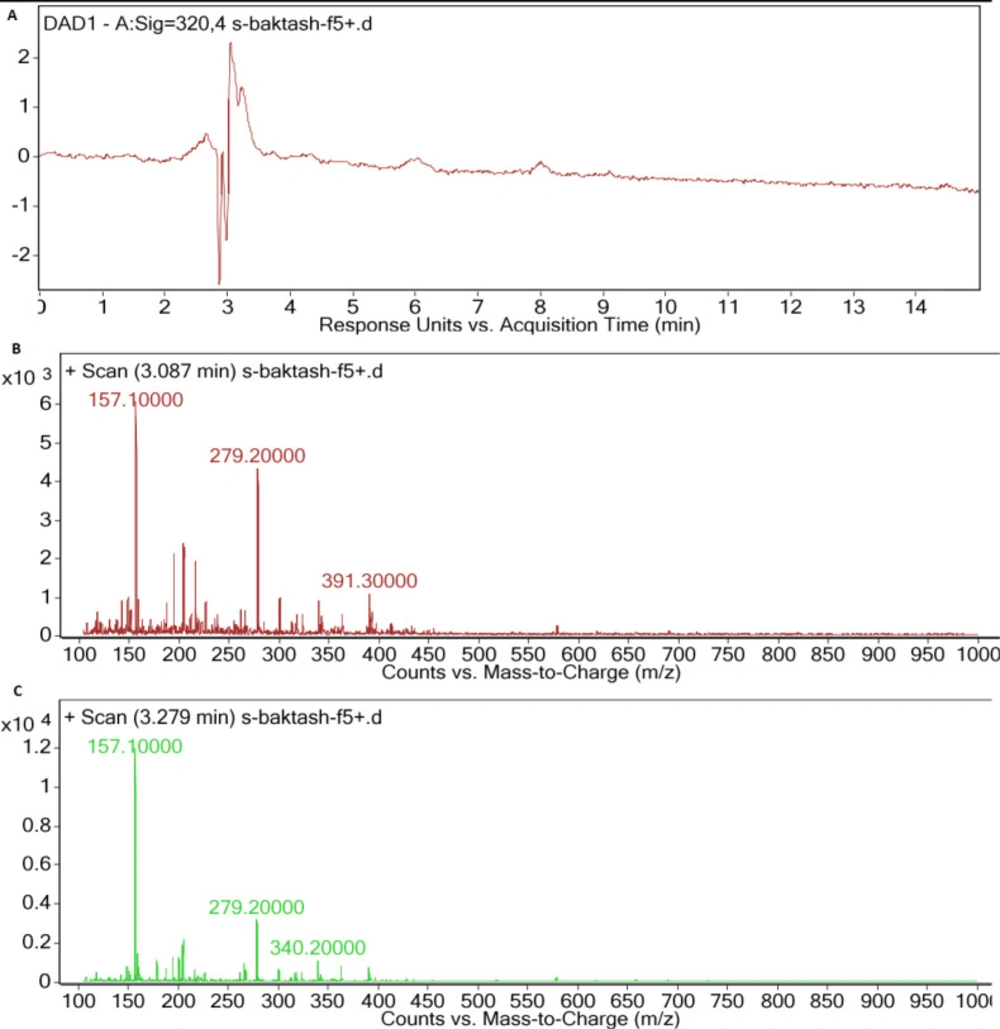
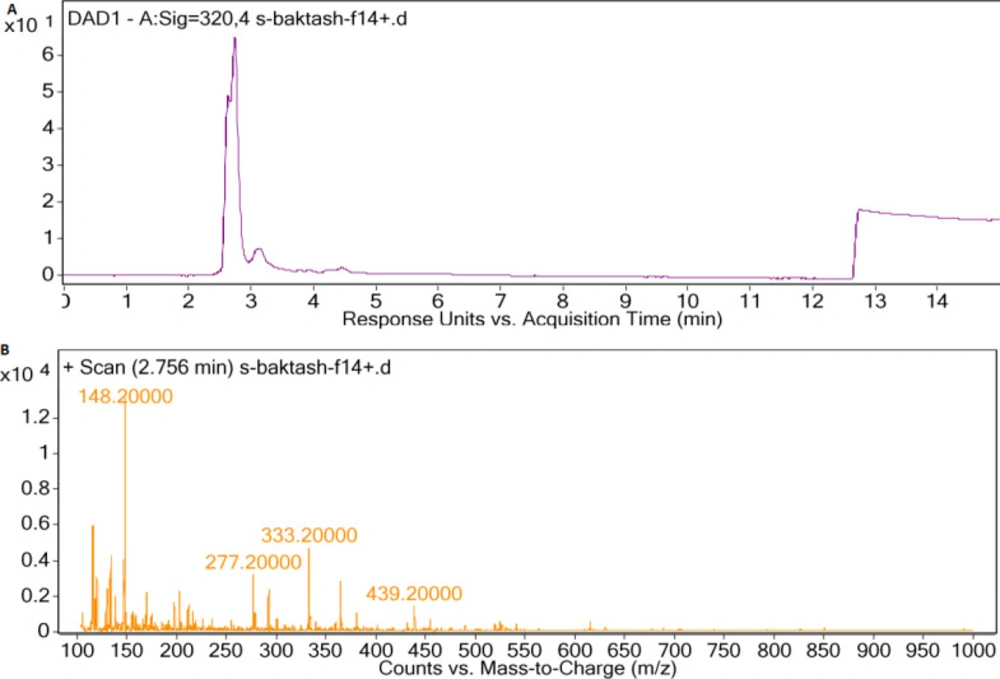
HPLC chromatogram of F5 extract displayed peaks at retention times of 3.08 and 3.27 min (Figure 5a), correlated with compounds having [M+H]+ ion peaks at m/z 391.3 (Figure 5b) and 340 (Figure 5c), respectively in the ESI-Mass spectra. Mass spectrum of F14 extract revealed an ion peak at m/z 333, correlated with compound detected at the retention time of 2.75 min in the HPLC chromatogram (Figure 6). Since the most reported MAAs had low molecular weights (< 400 Dalton) (21), these extracted mixtures might contain MAAs or their derivatives.
Discussion
Several UV-absorbing/screening MAAs have been reported from various organisms, including cyanobacteria (4). However, the production of MAA by different cyanobacteria species still needs to be investigated. In this study, we determined the production possibility of MAAs in two cyanobacteria species isolated from paddy fields of the north of Iran, Fischerella sp. (F5 and F14). Studies on MAAs production in Fischerella sp. were only limited to F. muscicola obtained from Thailand (13).
These isolated cyanobacteria had identified by the morphological study and then confirmed by16S rRNA PCR/sequencing analysis here. Methanol (100%) extraction as a common extraction method for partial purification of MAAs (22) was used in this study and then analyzed by HPLC and LC-MS methods. The results of HPLC analysis showed that both Fischerella species contained compounds with maximum absorption in the range of 300 to 400 nm. Therefore, it can be inferred that both species have the ability to produce secondary metabolites with MAA characteristics. According to the characteristics, like other MAAs, the extracted MAAs from these Fischerella sp. maybe contain a glycine moiety on the C3 and a second group on C1 of the cyclohexenimine ring. The addition of a second amino acid can result in MAAs like Porphyra-334, Shinorine, mycosporine-2-glycine, mycosporine-glycine-glutamic acid. In some MAAs like asterina-330. If and palythinol, the second group was amino alcohol, and in some others like palythene and usujirene, an enaminone system was linked to the C1, which resulted in maximum absorption at 360 357 nm, respectively. There were reported several compounds in which glycine has been replaced by methylamine such as mycosporine-methyl-amine-serine, mycosporine-methylamine-threonine (ʎmax = 325-330 nm) or to an amine group like palythine-serine, and palythine-threonine (ʎmax = 320 nm) (20).
According to the retention times of the components in HPLC chromatograms, it can be concluded, the extracted compound from F14 is probably more polar than those extracted from F5. However, the major peak of the HPLC chromatogram of F5 extract appeared at the retention time of 2.59 min and had a maximum absorption at about 290 nm, which may be related to the precursors involved in MAA production (23). The λmax of gadusol as the precursor of MAAs was reported at 269 nm (20). Pursuant to a number of studies, the use of stimuli such as UV-A and UV-B radiation, which induced MAA production, may increase the production of MAA-like compounds (24, 25). Therefore, in future studies, it will be considered to increase the MAAs production in these species by UV irradiation.
In the mass spectra of F5 species, ion peaks at 157, 279, and 391 m/z for one extracted compound, and 157, 279 and 340 m/z for another one were observed. Mass spectra and maximum absorption of the compounds extracted from F5 were in part similar to those of Porphyra (26), Mycosporine-Glycine-Valine (21), or their sugar derivatives.
In the mass spectra of the compounds extracted from F14 species, an ion peak at 333.2 m/z was observed, which is similar to that of Shinorine (21, 27 and 28). Similarly, Vanessa Geraldes et al. showed that out of the 69 cyanobacteria, which were isolated from Brazilian environments, 26 strains could synthesize MAAs. Among them, Fischerella sp. (CENA 161) produced Shinorine (29).
The species of F14 produced just one MAA compound; therefore, its purification and isolation may be easier and more cost-effective for using as a single active ingredient in sun-protective products even though extraction of the genes involved in the biosynthesis of Shinorine and its expression in other cyanobacteria can provide a valuable source for its production, same as what was previously performed by Yang et al. to develop engineered Synechocystis sp. PCC6803 for improved production of MAA (30).
However, further analysis is required to accurately identify these active metabolites of F5 and F14 Fischerella sp.
| Amplicon | Primer | Sequences | Tm | Site | Ref. |
|---|---|---|---|---|---|
| 630 | CYA106F | CGGACGGGTGAGTAACGCGTG | 65.8 | 106–127 | (16) |
| CYAN738R | GCTAGGACTACWGGGGTAT | 57.5 | 738–765 | ||
| 1260 | CYAN738F | ATACCCCWGTAGTCCTAGC | 57.5 | 738–765 | (17) |
| CYAN1281R | GCAATTACTAGCGATTCCTCC | 57.8 | 1281–1302 |
| Sample | Description | Max Score | Query Cover | Per. Ident | Accession |
|---|---|---|---|---|---|
| F5 | Fischerella sp. Sara 1 16S ribosomal RNA gene, partial sequence | 928 | 94% | 99.61% | KY618863.1 |
| Fischerella sp. MGCY374 16S ribosomal RNA gene, partial sequence | 928 | 94% | 99.61% | KY056817.1 | |
| F14 | Fischerella sp. ATCC 43239 16S ribosomal RNA gene, partial sequence | 627 | 95% | 86.52% | KJ768872.1 |
| Fischerella sp. IR-291 16S ribosomal RNA gene, partial sequence | 627 | 95% | 86.52% | KY011910.1 |
Conclusion
In this investigation, it was proved that both Fischerella species collected from the North of Iran are capable of producing MAAs, and therefore, these Fischerella species can be further studied and considered as an appropriate source of UV absorbing compounds. Since the culture of cyanobacteria is simple and low cost, they are an attractive source for the natural production of bioactive compounds. In addition, culture optimization and stimuli induction for improving the production of MAA compounds in future studies can result in obtaining a valuable source for the natural production of sun-protective agents. Due to the application of MAAs as valuable compounds in the cosmetics and pharmaceutical industries, further optimization studies in MAAs biosynthesis is highly attractive and important for investment.
Conflict to interest
The authors declare that there are no conflicts of interest.
Funding support
This work was financially supported by the Protein Technology Research Center of Shahid Beheshti University of Medical Sciences, Tehran, Iran (Grant# 16512; Ethical code: IR.SBMU.PHARMACY.REC.1398.037).