Introduction
Urinary tract infections (UTIs) are among the most frequent infectious diseases worldwide. UTIs comprise a range of disorders, including cystitis (infection of the bladder) and pyelonephritis (infection of the kidney) (1). Escherichia coli is the most predominant pathogen causing 80-90% of community-acquired UTIs and 30-50% of nosocomially-acquired UTIs (2). The development of antibiotic resistance has led to discovery of many natural mobile elements, including transposons and conjugative plasmids. Comparative sequence analysis of these elements has ultimately led to discovery of integrons (3) as elements which contain the genetic determinants of the components of a site-specific recombination system that recognizes and captures mobile gene cassettes (4). They are defined by the presence of an integrase gene (intI), a recombination site (attI), and one or two promoters responsible for expression of the inserted gene cassettes (5). Three classes of resistant integrons (RIs) have been defined on the basis of the divergence among their integrase genes, and each class appears to be able to acquire the same gene cassettes (3). Class 1 integrons are found extensively in clinical isolates, and most of the known antibiotic-resistance gene cassettes belong to this class. Class 2 integrons are exclusively associated with Tn7 derivatives and class 3 integrons are thought to be located in a transposon (6).
Integrons are gene exchange systems and are known to play a significant role in the acquisition and dissemination of antimicrobial resistance genes and to be selected by antimicrobial pressure. These systems play a broad and important role in MDR E. coli strains (strains showing antimicrobial resistance to multiple antimicrobial drugs) (5). There is a need for improved surveillance for drug resistance and its mechanisms of dissemination and persistence and mobility of resistance genes in the community and clinical settings.
Considering the role of E. coli in urinary tract infections and increasing antibiotic resistance in this bacterium on the one hand and the importance of integrons in acquiring antibiotic-resistant gene cassettes on the other hand, the aim of this study was to investigate the antibiotic resistance and the frequency of class 1 and 2 integrons in E. coli strains isolated from urinary tract infections.
Experimental
Sample collection and antibiogram
In this cross-sectional study, that was conducted over a period of six months from January 2016 to June 2016, a total of 100 E. coli isolates were collected from UTI samples of outpatients referring to Baqiyatallah Hospital, Tehran, Iran. UTI in patients was confirmed based on clinical signs and laboratory findings related to this infection. All strains were identified using conventional biochemical tests. The bacterial suspension was stored in TBS medium containing 20% glycerol at -70 °C. The Antibiotic susceptibility pattern of strains was determined using disk diffusion method (Kirby-Bauer) on Mueller Hinton agar medium (Merck, Germany) according to the CLSI guidelines (7). The antibiotics disks used in this study were as follows: Ampicillin, amikacin, gentamicin, ceftazidime, cefotaxime, ciprofloxacin, co-trimoxazole, and nalidixic acid.
DNA extraction and PCR
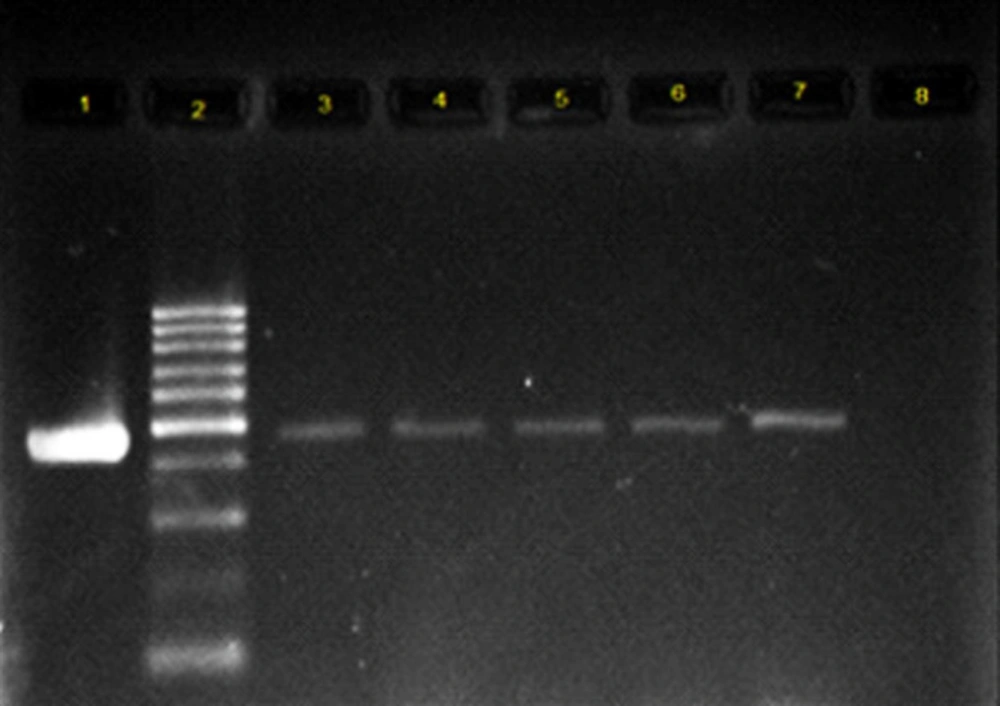
All the collected isolates were extracted using DNA extraction kit, a product of Bioneer Co. of Korea, according to the manufacturer’s instructions. PCR test was conducted to identify the genes coding for integrase enzyme. The reaction was done in a total volume of 20 μL, so that 1 μL of each primer, 1 μL of DNA template, 10 μL of Master Mix and 7 μL of distilled water were used. Primers used are shown in Table 1 (8, 9). Class 1 integron amplification was done in the following conditions: denaturation temperature of 95 °C for 5 min, 30 thermal cycles with denaturation temperature of 94 °C for 50 seconds, Primer binding temperature of 54 °C for 50 seconds, amplification temperature of 72 °C for 1 minute, and final amplification temperature of 72 °C for 5 min. Class 2 integron amplification conditions were similar to those of class 1 integron gene, but the annealing temperature was 52 °C. Electrophoresis of PCR products was performed on 1.5% agarose gel for 1 h in voltage of 80 volts. Then, they were stained using ethidium bromide. The results were observed using Gel Documentation system and examined under ultraviolet light. To validate the test, the confirmed control strains of E. coli, containing the integron gene were used as the positive control and the microtubes, containing the reaction material with no DNA template were used as PCR test contamination control (Figure 1 and 2). The PCR product sequencing was done by Bioneer Co. of Korea as ordered by Takapouzist Co. in Tehran.
Statistical analysis
The data were analyzed using SPSS statistical software, version 22 (SPSS Inc., Chicago, IL). The association between presence of class 1 integron and antibiotic resistance was determined by Mann-whittney test. A p-value of > 0.05 was considered statistically significant.
Results
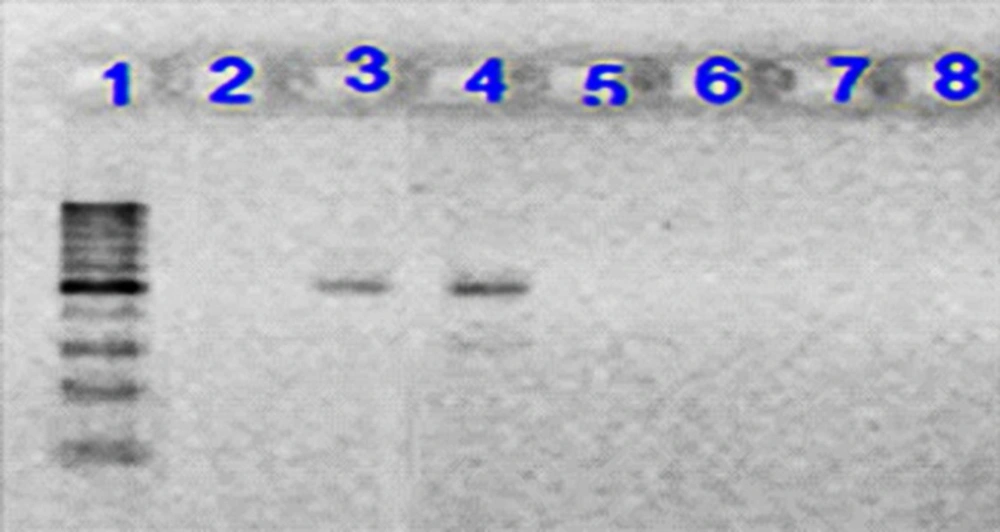
The most frequent antibiotic resistance was observed to ampicillin (72%), co-trimoxazole (66%), and nalidixic acid (62%). The highest sensivity was seen to amikacin (11%) and gentamicin (20%). All strains were resistant to at least one antibiotic and MDR strains that were observed in 80% of E.coli isolates. Molecular analysis for the presence of class 1 and 2 integrons showed that 70% of the isolates had class 1 integron, while class 2 integron was observed only in 3% of the strains. The results of Mann-whittney test showed the statistically significant relationship between the presence of class 1 integron and resistance to ampicillin, gentamicin, ciprofloxacin, co-trimoxazole, and nalidixic acid. There was no significant association between the presence of class 1 integron and the resistance to amikacin, ceftazidime, and cefotaxime (Table 2).
Discussion
The level of antibiotic resistance among hospital and community-acquired isolates has steadily increased and become a major global health problem. Antibiotic resistance pattern of isolates from UTI, which is one of the most frequent infectious diseases and most common infection in hospital and care institution, is also changing (10). In the present study, which was conducted on E. coli strains causing urinary tract infections, the most susceptibility was observed to ampicillin, co-trimoxazole, and nalidixic acid. The antibiotic resistance to ampicillin in Jahrom, Hamedan and in similar studies in Turkey, India, Taiwan, and Mexico has been reported as 80.2%, 61.9%, 73.3%, 90%, 82.9%, and 83.7%, respectively (11-16). After the ampicillin, co-trimoxazole was the second ranked antibiotic in terms of showing the antibiotic resistance as follows: Jahrom: 76%, Hamedan: 56.4%, and Turkey: 63.3%, which was similar to the present study (11, 13, 16). In the present study, the most effective antibiotics against E.coli isolates were amikacin and gentamicin so that only 11% and 20% isolates were resistant to amikacin and gentamicin, respectively. The resistance rate to amikacin and gentamicin was as follows: in Hamadan, 6.1% and 18.8%; in Turkey, 4.9% and 13.9%; and in Mexico, 1.7% and 23.9% (13, 14, 16).
In the present study, MDR strains were observed in 80% of E.coli isolates. The level of antibiotic resistance among E. coli strains causing urinary tract infection varies from country to country. For example, in the United States (2000), India (2012), and Korea (2015), the frequency of MDR strains was reported as 7.1%, 75%, and 21.9%, respectively (17-19). In recent years, a high percentage of MDR E. coli strains have been reported in different parts of Iran (11, 20-23). The aim of the present study was to investigate the role of integrons in antibiotic resistance, which plays a broad and important role in MDR E. coli strains. In the present study, class 1 and 2 integrons of E.coli isolates were detected in 70% and 3% of isolates, respectively. Previous studies in the other parts of the world also investigated the frequency of different classes of integrons. In a study conducted in 2008 by Farshad et al., 6.25% and 10.41% of strains had class 1 and class 2 integrons, respectively (24). In 2011, Rezaee et al. investigated the prevalence rate of class 1 and 2 integrons among 140 clinical isolates of E. coli. Their results showed that 22.5% and 5.08% of the isolates had class 1 and class 2 integrons, respectively (25). In a study conducted in 2013 by Ranjbaran et al., class 1 and class 2 integrons were detected in 86% and 8% of the isolates, respectively (26). In a study by Khorramrooz et al., the frequency of class 1 and 2 integrons was 52% and 2.5%, respectively (22). In previous studies in California, Pakistan and Syria, the frequency of class 1 integron were reported as 49%, 43.56%, and 54.6%, respectively (27-29). In a study in 2015, Lin et al. showed that 25 isolates of 162 isolates had class 1 integron. No class 2 integron was found in Malaysia (30).
One of the limitations in the present study was that we were unable to determine gene cassettes carried in the intergrons coding the resistance to the mentioned antibiotics. However, our data indicate that there is a significant correlation between class 1 integrons and resistance to ampicillin, gentamicin, ciprofloxacin, co-trimoxazole, as well as nalidixic acid. The resistance to these antibiotics could be due to the presence of resistance gene cassettes in this class of integrons. In the cases where there was no significant correlation between the presence of class 1 integrons and the antibiotic resistance, the resulting resistance can be achieved through other ways such as the presence of the resistance genes on resistance plasmids and etc. It is recommended to conduct more detailed studies on the nature of integrons, the gene cassettes carried in the intergrons, and the other resistance mechanisms in these species to provide better therapeutics and control strategies in the future.
| Reference | Amplicon Size (bp) | Primer Sequence | Gene |
|---|---|---|---|
| (8) | 484 | 3'-GGGTCAAGGATCTGGATTTC –F: 5' | Int1 |
| (9) | 466 | GCAAATGAAGTGCAACGC-3'–F: 5' | Int2 |
| Antibiotics | Isolated having integrons (n = 70) | Isolates lacking integrons (n = 30) | p-value* | ||||
|---|---|---|---|---|---|---|---|
| Susceptible NO (%) | Intermediate NO(%) | Resistant NO(%) | Susceptible NO (%) | Intermediate NO(%) | Resistant NO (%) | ||
| Ampicillin | 7 (10%) | 2 (2.58%) | 61 (87.14%) | 15 (50%) | 4 (13.33%) | 11(36.66%) | <0.001 |
| Amikacin | 50 (71.42%) | 14 (20%) | 6 (8.51%) | 19 (63.33%) | 6 (20%) | 5 (16.66%) | 0.4 |
| Ceftazidime | 32 (45.72%) | 5 (7.14%) | 33 (47.14%) | 16 (53.33%) | (0%) | 14 (46.66%) | 0.5 |
| Gentamicin | 56 (80%) | 2 (2.58%) | 12 (17.14%) | 15 (50%) | 7 (23.33%) | 8 (26.66%) | 0.008 |
| Ciprofloxacin | 24 (34.29%) | 1 (1.42%) | 45 (64.29%) | 21 (70%) | 4 (13.33) | 5 (16.66%) | <0.001 |
| Co-trimoxazole | 11 (15.71%) | 0 (0%) | 59 (84.28%) | 18 (60%) | 5 (16.66%) | 7 (23.33%) | <0.001 |
| Nalidixic acid | 17 (24.28%) | 3 (4.28%) | 50 (71.42%) | 14 (46.66%) | 4 (13.33%) | 12 (40%) | 0.005 |
| Cefotaxime | 28 (40%) | 0 (0%) | 42 (60%) | 15 (50%) | 0 (0%) | 15 (50%) | 0.3 |
Electrophoresis of PCR product on 1.5% agarose gel for Int2 gene. Lane 1: 100 bp DNA ladder as the molecular size marker; Lane 2: PCR mix with no template (negative control); Lane 3: positive control for Int2 gene; Lane 4: the Int2 gene detected in UPEC strain; Lane 5-8: UPEC strains negative for Int2 gene