Introduction
Ciprofloxacin is one of the most widely-used antibiotics for the treatment of infections, such as urinary tract infections, caused by Gram-negative bacteria, like E. coli (1, 2). However, there is growing evidence which shows that the frequency of resistance to ciprofloxacin among E. coli isolates is rising yearly all over the world (3, 4, 5). This resistance has been linked to the chromosomal mutations that cause either changes in gyrA and gyrB, the main target of antibiotic or decreases the accumulation of drug inside the bacteria. However, in higher quinolone-resistant mutants, both of these are the case (6).
In the previous study, we described E. coli ciprofloxacin resistant mutants that have mutations in gyrA gene (7). This gene, along with gyrB gene, encodes DNA gyrase subunits (7, 8). This enzyme catalyzes the negative supercoiling of DNA required for chromosome replication, transcription and recombination (8, 9).
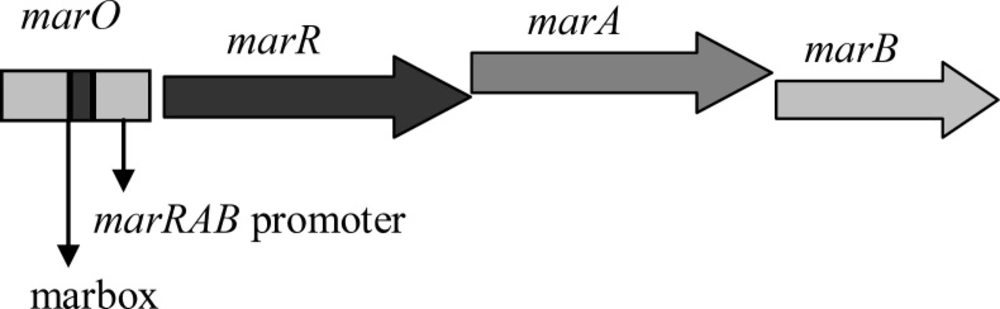
Schematic representation of marRAB operon in E. coli. marO harboring promoter and marbox. Simplified and adapted from Cohen et al., 1993 (14).
These gyrA mutants have different MICs against ciprofloxacin (7). Thus, it is possible that some with higher level of resistance may have extra mutations that decrease the amount of ciprofloxacin inside the cells (6). This may result from the activation of endogenous transmembrane efflux pump, AcrAB-TolC (10). This is a multidrug efflux pump consists of three components, including AcrB, the inner membrane transporter protein, TolC, the outer membrane channel, and AcrA, the periplasmic membrane fusion protein (10). This pump is activated through MarA. MarA is a transcriptional activator which activates its own transcription and that of a large number of mar regulon genes, including AcrAB, tolC and micF, the repressor of OmpF, via binding to special sequences called marboxes that are located in the upstream of promoters for the target genes (11, 12). This protein is produced by marA, a member of marRAB operon (Figure 1). MarR is another member of operon which encodes a repressor protein. MarR controls the intracellular levels of MarA through binding to two certain sequences in marO, the regulatory region of operon. One is between -35 and -10 sequences and the other is just near the initiation codon (13). Thus, mutations that inactivate marR can increase the expression of marA gene and thereby can enhance the ejection of ciprofloxacin and decrease the entry of some other antibiotics, such as ampicillin, chloramphenicol, and tetracycline. This happens first, through the increase in the activity of AcrAB-TolC pump and second, via the decreases in the synthesis of porin proteins, OmpF, which are used as a route of cell entry for above mentioned antibiotics. It was proposed that mar regulon is associated with clinical antibiotic resistance and treatment failure (14). This regulon is also related to the organic solvent tolerance. The correlation between the high levels of organic solvent tolerance and low levels of resistance to some antibiotics as mentioned above has already been explained (15, 16).
| Strain/mutant | MIC (ng/mL) |
|---|---|
| MG1655 wild type | 35 |
| W10-W11 | 62.5 |
| W25-W26 | 75 |
| W44-W45 | 125 |
| W47-W48 | 312 |
| W49-W50 | 625 |
It was reported that some of the quinolone-resistant E. coli isolations acquired constitutive expression of marRAB operon due to the occurrence of mutations in marR (17). To gain more information on extra mechanisms of resistance in gyrA mutants, they were examined for acquirement of high organic solvent tolerance, resistance to tetracycline and mutations in marR gene.
Experimental
Antimicrobial agent and chemicals
Tetracycline hydrochloride (Tc) was obtained from Sigma. Stock solution was 4 mg/mL. Organic solvents used for this study were n-hexane (Merck) and cyclohexane (Merck).
Bacterial strain and mutants
MG1655 was parent strain. GyrA mutants isolated in previous work (7) are listed in Table 1.
Media
LB broth (Merck) was used to prepare LBGMg agar medium, containing 0.1% glucose, 10 mM MgSO4 and 1.5% agar other than LB.
Organic solvent tolerance assay
Serial dilutions were prepared from fresh cultures of strains and mutants in 0.9% NaCl and 5 µL of each dilution spotted on a solid LBGMg medium as described previously (15). The surface of the medium was overlaid with an organic solvent and incubated at 37°C for 24 h. Then, the number of colonies per spot was counted on each plate.
| Stran/mutant | No. of bacteria spotted | ||||
|---|---|---|---|---|---|
| Without solvent | Ha | H-CHb (3:1) | H-CHb (1:1) | H-CHb (1:3) | |
| MG1655 | 30*106 | 19*104 | 0 | 0 | 0 |
| W10-W11 | 26*106 | 19*104 | 0 | 0 | 0 |
| W25-W26 | 22*106 | 6*105 | 0 | 0 | 0 |
| W44-W45 | 20*106 | 19*104 | 0 | 0 | 0 |
| W47-W48 | 20*106 | 21*104 | 0 | 0 | 0 |
| W49 | 17*106 | 5*105 | 0 | 0 | 0 |
| W50 | 19*106 | 21*104 | 0 | 0 | 0 |
Antibiotic susceptibility test
As described in previous study (7), MICs of Tc for control strain, MG1655, and gyrA mutants were determined using broth dilution method (18). Different concentrations of Tc ranging from 0.125 µg/mL to 5 µg/mL, were used. MICs for control strain and gyrA mutants were determined in three independent experiments.
PCR amplification and DNA sequencing
A single colony from each strain and mutants grown on LB agar was used as a template for PCR reaction as described previously (7). Primers used for PCR amplification and DNA sequencing were forward primer 5`-GGTGGTTGTTATCCTGTGTA-3` and reverse primer 5`-CGGCAGGACTTTCTTAAGC-3`. PCR products (700 bp in size) which contained part of marO and the entire marR gene were sequenced.
Results and Discussion
Ten gyrA mutants and their parent strain MG1655 were assessed for organic solvent tolerance. Results are shown in Table 2. As can be seen from this table, MG1655 can grow in presence of hexane, but not cyclohexane. This is consistent with the previous result for this strain (15). All mutants derived from this strain also show nearly the same results. Some mutants had slightly better growth on hexane.
To assess whether gyrA mutants may acquire resistance to other antibiotics, the MIC of Tc for control strain and gyrA mutants were measured. For MG1655, MIC was 3 µg/mL (Table 2). This is consistent with previous data (19). MICs of gyrA mutants were nearly the same as that of MG1655, except some seemed to show better growth at 3 µg/mL tetracycline and their MICs were 4 µg/mL (Table 2). Collectively, these results suggest that gyrA mutants may either not acquire a mutation in marR gene, or some of them acquire a mutation, but cannot cause high level of organic solvent tolerance and partial resistance to other antibiotics at once and need to be induced in the presence of inducers, such as tetracycline (19).
| Strain/mutant | MIC* (μg/mL) |
|---|---|
| MG1655 wild type | 3 |
| W10-W11 | 3 |
| W25-W26 | 4 |
| W44-W45 | 3 |
| W47-W48 | 3 |
| W49 | 4 |
| W50 | 3 |
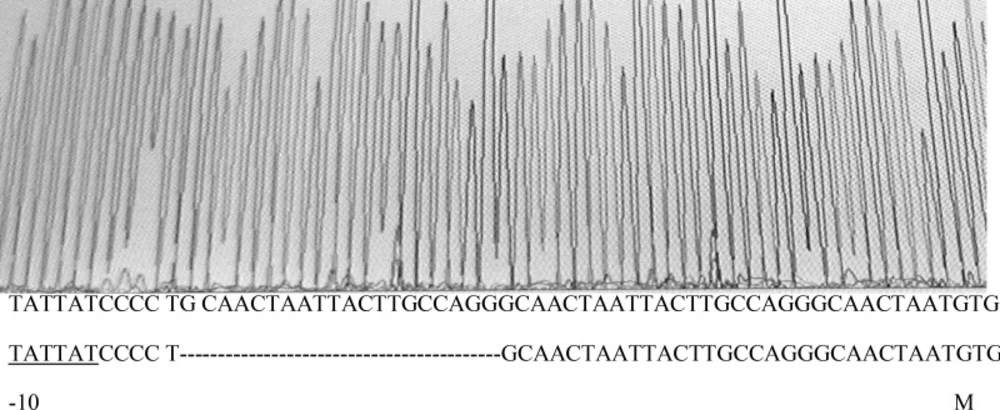
To verify this marR gene and its upstream region, marO in MG1655 and its 10 gyrA mutants were amplified. Figure 2 shows the result of gel electrophoresis of the marOR PCR product of MG1655. The same results were obtained for all gyrA mutants. Then, the PCR products were sequenced using forward and reverse primer. Finally, the sequences were compared with the published marR sequence of MG1655 strain, using EBI bioinformatics program for nucleotide pair wise alignment. MG1655 showed the complete match with previous published sequence for marR (data not shown). Moreover, 7 gyrA mutants also showed the same result as MG1655. Their Marbox and promoter site were also intact. This suggests that their higher ciprofloxacin MICs may not be associated to the induction of drug efflux. However, 2 mutants with higher Tetracycline MIC, had mutations in marOR. One contained a T→C change at nucleotide position 221 in coding region of marR that alters methionine-74 to threonine (Figure 3), but its marbox and promoter site was without change. The other mutant harbors, a 20-base pair tandem duplication of a sequence (GCAACTAATTACTTGCCAGG) started 6-base pair downstream from the 10-position of promoter site (Figure 4). This type of mutation was described before (14). However, its marbox was intact. The last mutant with higher tetracycline MIC did not possess a mutation in marOR. This suggests that the reason of slightly different tetracycline MIC may be the occurrence of mutation in a gene that encodes a suppressor for the pump.
In the previous study, ciprofloxacin-resistant mutants which possess mutations in gyrA were isolated (7). These gyrA mutants had different levels of resistance to ciprofloxacin. It was explained that one reason for this difference is due to the decreased levels of drug accumulation (6). The intracellular amount of antibiotic can be regulated through a efflux pump, AcrAB-TolC (10). The activation of this pump is associated with the presence of MarA, the transcription activator protein. The gene that encoded this protein is located in marRAB operon, consisting marR, marA and marB genes. This operon is normally inactivated through the product of marR gene, MarR. The inactivation of MarR is caused through the mutations in marR (17). This leads to the expression of a multidrug resistance phenotype (MDR). This phenotype is also associated to the high level of organic solvent tolerance (15, 16). Therefore, in this study, ciprofloxacin-resistant mutants with different MICs were examined for the tolerance of organic solvents, resistance to tetracycline and presence of marR mutation.
The obtained results revealed that none of the gyrA mutants even those with higher ciprofloxacin MICs possess the MDR phenotype de novo, but some gyrA mutants may gain this phenotype gradually following the exposure to an inducer of marRAB operon, such as tetracycline. This was revealed through the discovery that 3 out of 10 gyrA mutants are slightly more tolerant to hexane and resistant to tetracycline compared to MG1655.
Our finding that 7 gyrA mutants are the same as MG1655 for sensitivity to tetracycline, tolerance of organic solvent and devoid of marOR mutation, implies that the reason for their higher ciprofloxacin MICs in comparison with MG1655 may be the presence of mutations in gyrB, parC or parE (6). The last two genes encode the subunits of topoisomerase IV, the minor target of fluoroquinolone antibiotics in E. coli (6).
Moreover, an E. coli mutant (OST3408) was found by Asako that is tolerant not only to hexane, but also to cycloheexane (15). Most of E. coli strains are sensitive to cyclohexane. This mutant contains a substitution of serine for arginine at position 73 in the coding region of marR. The MarR protein has been shown to contain the helix-turn-helix motif started from the 60th amino acid codon and extended to the 80th one (20). This motif is necessary for the binding of repressor to the promoter site. However, we found that a T→C mutation in marR that causes a substitution of threonine for metionine at position 74 does not confer tolerance to cyclohexane. This implies that both the position of amino acid and the type of substitutions in amino acid sequence of this motif are important on functionality of repressor. On the other hand, the alteration of the repressor binding sites interferes with repressor activity. We found a marR mutation with altered repressor binding sites that has already been discovered (14). It was found that this kind of mutation increases the activity of marRAB operon (14).
Moreover, mutations in acrR gene, encoding the repressor of AcrAB-TolC pump called AcrR, increase the activity of the pump (21). This fact that one of the three slightly resistant mutants did not possess a mutation in marOR, implies that this gyrA mutant may acquire a mutation in acrR gene.
Furthermore, highly organic solvent tolerant mutants gain mutations both in marOR and acrR (22). Therefore, it is possible that our gyrA marR double mutants are not resistant to cyclohexane due to their lack of a mutation in acrR.