Introduction
Streptomyces clavuligerus is an actinomycet, which produces more than 21 bioactive secondary metabolites (1-3). Between them, metabolites with β-lactam ring such as cephamycyn C, penicillin N and cephalosponins with antibacterial effect, and clavulanic acid, a β-lactamase inhibitor, are more important (4). Clavulanic acid is a β-lactam antibiotic with weak antibacterial effect and strong β-lactamase inhibition (4-6). Augmentin®, a brand name, containing a combination of amoxicillin and potassium clavulanate, is one of the best selling antibiotics (7-15). Clavulanic acid is produced industrially by fermentation of Streptomyces clavuligerus and researches have increased its concentration (up to ≈1.4 g L-1) by strain improvement, recombinant DNA technology, and media composition and growth conditions optimization (3, 7, 10, 15-24). UV irradiation is one of the strain improvement strategies through random mutation (7, 16). UV radiation, in the range of 200-300 nm, produces thymidine dimmers and increases probability of deletion during the duplication process (25, 26). UV is a very convenient and relatively safe mutagen (27), however very few researchers (7, 16) have used this technique to overproduce clavulanic acid. The main objective of this study was to increase the level of clavulanic acid production by S. clavuligerus, using UV irradiation.
Experimental
Microorganism and cultivation
Streptomyces clavuligerus (DSM 738) was revitalized on GYM- broth and maintained on GYM-Agar. Sodium chloride solution (0.9%) containing tween-80 (0.2%) was poured onto GYM-Agar medium, on which S. clavuligerus was grown for 7 days at 28 °C. The spores and aerial mycelia were scraped off the agar surface, using a sterile loop. After filtering through cotton wool, the spore suspension was serially diluted to 102 CFU mL-1 and one mL of this suspension was poured and spread onto each sterile glass plate containing GYM-Agar and caffeine (0.5 g L-1) (28).
UV Irradiation
The plates were irradiated with UV light (200-300 nm, 1.7 W m-2) for 20 min, at a distance of 8 cm. After the irradiation, in order to protect against photoreactivation, the plates were wrapped with aluminum foil and incubated for 7 days. Finally, the colonies were sub-cultured (7 days, 28 °C) again to express and stabilize the mutants.
Bioassay
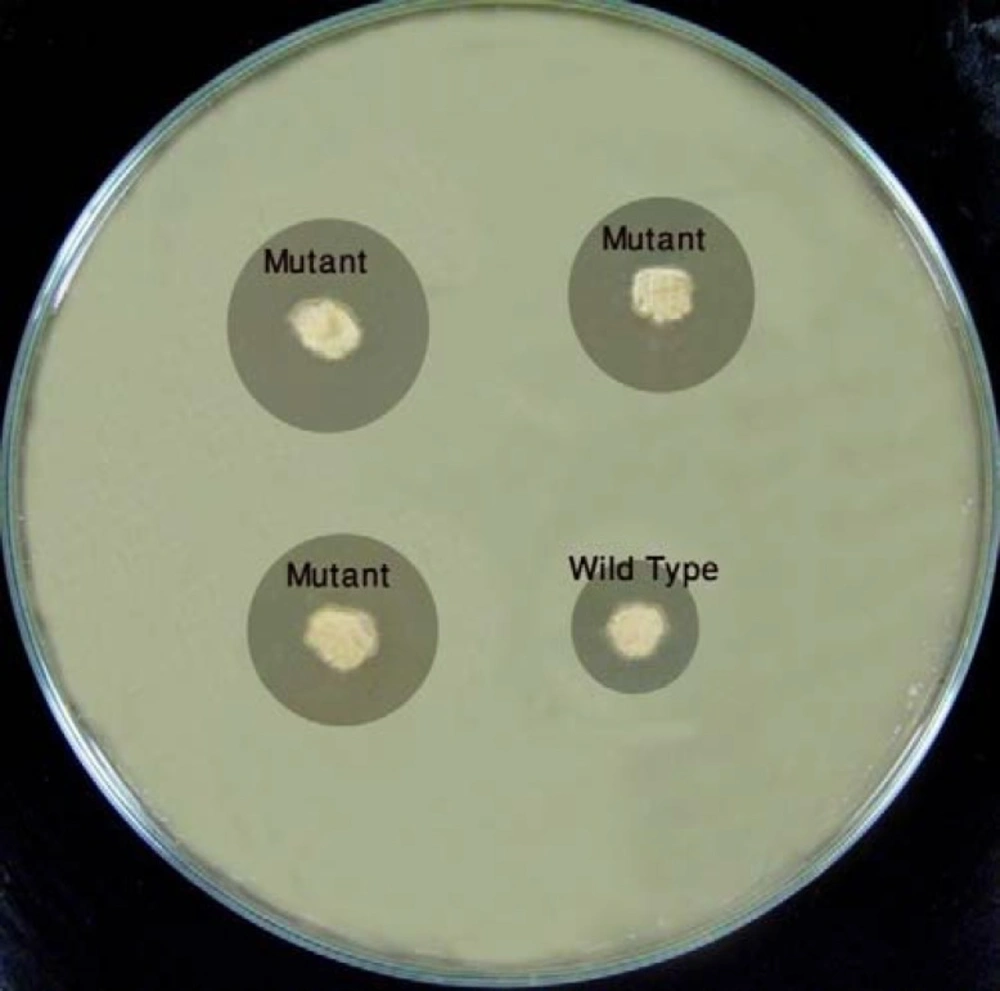
A radically modified bioassay method, based on the method developed by Lee et al (7), was used. Tenμµl of the spore suspension (102 CFU mL-1), prepared from the resulted culture, was poured into each of the 3 wells (out of 4 wells) in GYM- agar plates. A spore suspension (102 CFU mL-1) of the wild type micro-organism was poured into the 4th well. After an incubation period of 6 days at 28 °C, the plates were overlaid with nutrient-agar containing penicillin G (7 µg mL-1) and Klebsiela pneumoniae (105 CFU mL-1), and at the end again incubated for 24 h at 37 °C. The inhibition zone diameter was measured and compared with the wild type colony.
Clavulanic acid production
The pH of the seed medium (20 mL) consisting of peptone (1%), malt (1%) and glycerol (2%) was adjusted to 7.0 ± 0.1. The medium was inoculated (5%) and incubated (28 °C, 24 h, 220 rpm) in a 100 mL shaking flask. The production medium (50 mL) containing malt extract (3%), glycerol (0.5%), soy flour (0.5%), FeSO4 (0.0001%), and MnSO4 (0.0001%) was inoculated (5%) with the seed culture and incubated (28 °C, 220 rpm) in a 250 mL shaking flask.
HPLC analysis
Clavulanic acid was quantified by HPLC, using a reversed-phase column (C-18). The mobile phase contained KH2PO4 (50 mM, 70%, 0.348 mL min-1) and methanol (30%, 0.157 mL min-1) and the pH was adjusted to 3.2. Samples were derivatized using imidazole. The derivatives were detected at 311 nm (29).
Statistical analysis
Statistical analyses were performed through the Mann-Whitney non-parametric test, using the statistical package SPSS for Windows version 13.0 software (SPSS Inc, Illinois).
Results
During the subculturing of S. clavuligerus on GYM agar for sporulation, it was observed that by using deep (≈ 8 mm) agar plates denser colonies appeared with more grey pigments. Pale yellow droplets were secreted 6-7 days after the growth, which disappeared after leaving holes on the surface of S. clavuligerus colonies for a period of 24 h. This procedure has not been described before.
It seems that a thicker layer of agar in the plates, resulting in a greater population of the microorganism, could cause a higher and faster production of secondary metabolites, possibly containing antibiotics and clavulanic acid. After a while, these droplets might have been diffused in the plates.
Development of a bioassay for clavulanic acid
In order to screen overproducing mutants, a cheap, quick and fairly reliable bioassay method considering S. clavuligerus as clavulanic acid producer, and K. pneumonia as the sensitive microorganism was needed. When the bioassay was performed about 4 days after the growth of S. clavuligerus, not only the inhibition zone of K. pneumonia was not observed, but also a significant overgrowth zone was seen. However, when this assay was performed after 6 days, the inhibition zone was clearly observed (Figure 1).
It might be interpreted that some primary metabolites stimulating the growth are secreted by S. clavuligerus colonies during 4 days, which can lead to a faster growth of K. pneumonia. On the other hand, after 6 days, secondary metabolites (such as clavulanic acid), inhibiting the growth, might have been produced.
Screening of overproducing mutants
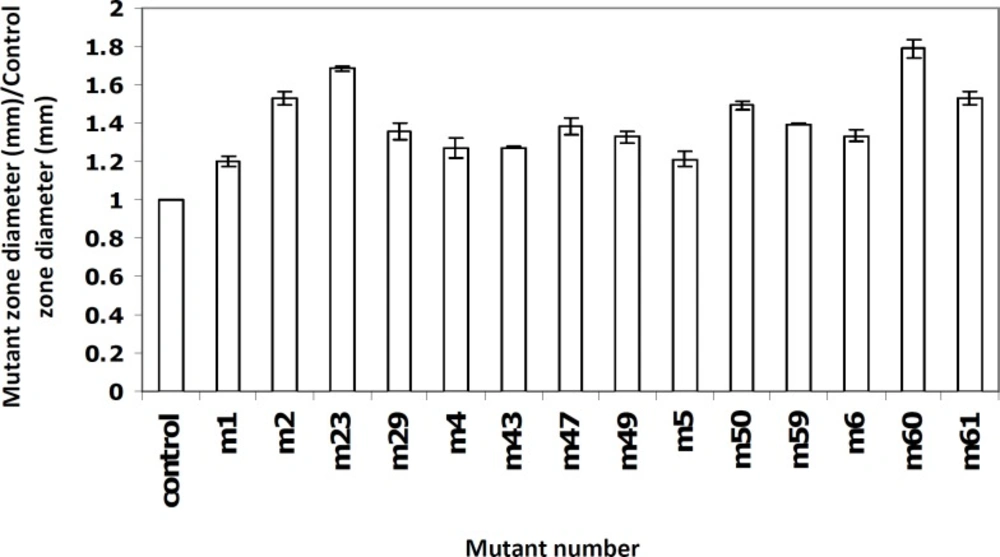
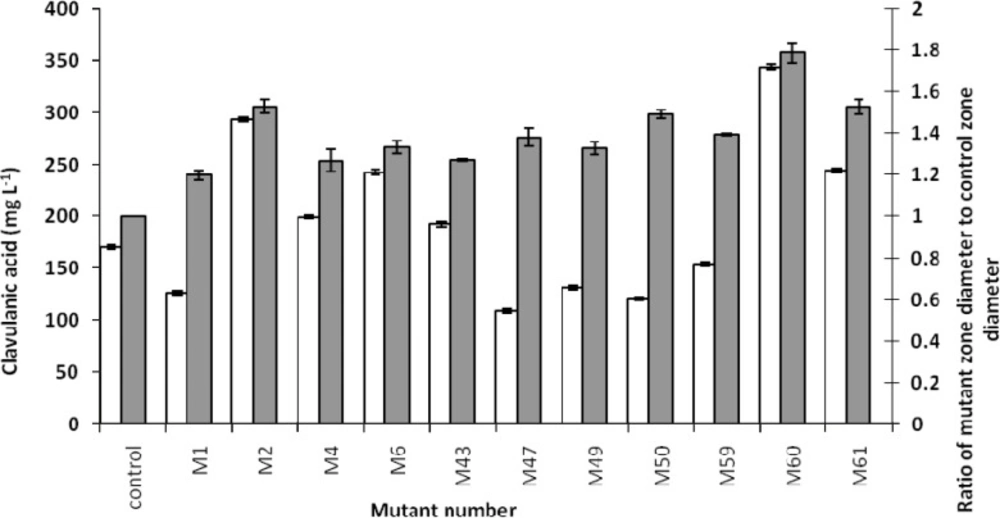
As mentioned earlier, a bioassay method was used to screen the overproducer mutants. Fourteen (out of 105) mutants showed inhibition zones more than the wild type control, with higher values for the mutants M60, M23 and M61 (Figure 2). Based on the Mann-Whitney statistical test, the differences between groups were found to be significant (P < 0.001).
Clavulanic acid determination by HPLC
The bioassay method used did not prove the production of clavulanic acid by the screened mutants. Therefore, we needed to confirm this and determine clavulanic acid concentration. Overproduction of clavulanic acid by the screened mutants was verified by the HPLC measurements (Figures 3 and 4). Intra-day and inter-day variations in precision (CV %) and accuracy (Error %) were found to be acceptable.
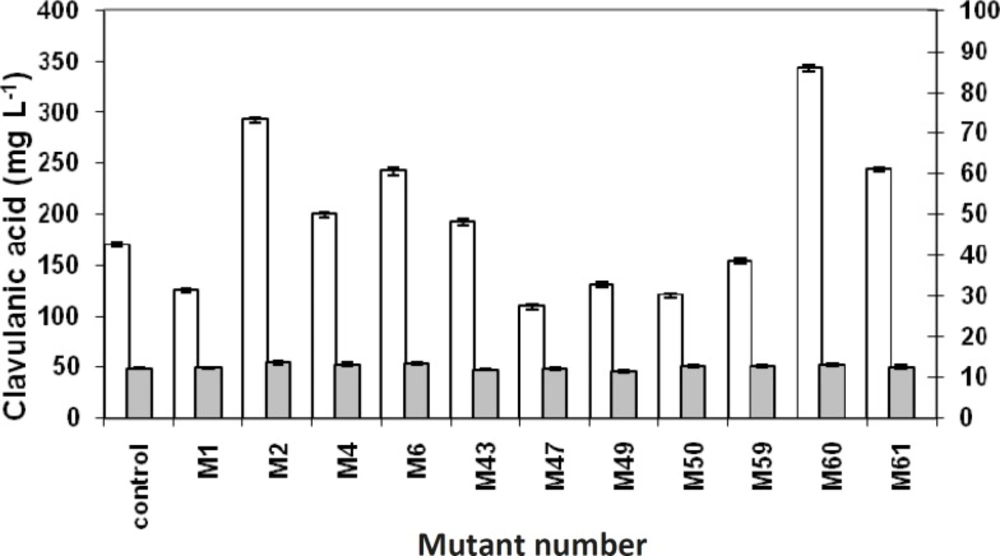
Comparing the concentrations of produced clavulanic acid with the biomasses of related mutants, clavulanic acid concentrations were found to be different among the various mutants. However, there was no considerable difference in the biomass concentrations (Figure 4). It might be interpreted that the overproduction of clavulanic acid, following the mutations, is not the result of overgrowth. This could be due to the changes in the metabolic pathway of the microorganism, resulting in the overproduction of clavulanic acid There was an analogy between the bioassay and HPLC method (Fig. 5), except in few cases, which might be due to the overproduction of other growth limiting factors.
Clavulanic acid production versus the ratio of mutant zone diameter to control zone diameter. Clavulanic acid concentrations (white columns) were measured by HPLC, and the ratio of mutant zone diameter to control zone diameter (gray columns) was measured by Vernier caliper and division. Data points have been expressed as mean ± standard error (n = 3).
Productivity of the selected mutants was also calculated (Table 1).
| Mutant number | Clavulanic acid concentration | Production time | Productivity |
|---|---|---|---|
| M2 | 293.11 | 72 | 4.07 |
| M6 | 242.67 | 72 | 3.37 |
| M60 | 343.87 | 72 | 4.77 |
| M61 | 244.38 | 72 | 3.39 |
Productivity of four selected mutants, investigated in this study
Discussion
The production of clavulanic acid was successfully increased 1.8 times by UV mutagenesis. It seems that the mutations occurred in the genes which were involved in the metabolic pathway of clavulanic acid production, but not in the primary metabolites responsible for growth. These mutants can be used industrially for production of clavulanic acid. A group of researches (7) increased clavulanic acid production up to 8 folds by UV mutagenesis and medium optimization. We increased it up to 1.8 folds, using random mutagenesis, which can be continued more considering the fact that “mutation is an accidental event”. In another study (30), we also inserted claR gene in E. coli XLI blue, using PZSclaR plasmid, as a vector to increase clavulanic acid production. In the near future we will combine these methods, and by using the medium optimization, it is hoped to increase the productivity.