1. Background
Microbial proteases show various applications in different industries (1). They are used as therapeutic agents and have an important place in the pharmaceutical industry (2). The microbial protease enzymes are used as inflammation inhibitors, anticancer medicines, thrombolytic agents, and anticoagulants (3). In addition, they serve as reagents in wound debridement to remove dead cells and treat damaged tissues (4). Microbial proteases were used in the injured spinal cord to remove scars and chondroitin sulfate to grow axons again (5).
Trypsin is a serine protease enzyme extracted from the bovine or porcine pancreas due to its activity on cell membrane proteins used in cell culture. The digestion of proteins in the adhesion site of cells to the culture vessels resulted in the detachment of cells (6, 7). The degradation of these surface proteins and the detachment of cells altered the cell morphology and metabolic pathways (8, 9). The trypsin treatment induces the round shape cells with new physiological activity (8). After separating the cells, trypsin should be neutralized by fetal bovine serum (FBS). In this reaction, FBS’s alpha-1 anti-trypsin binds to the trypsin and deactivates its protease activity (10). Trypsin is available either in lyophilized powder or in a solution form. The large-scale cell cultures are used to produce vaccines, recombinant proteins, and skin cell cultures. Due to unwanted immune reactions or contamination by pathogen reagents, animal-origin proteins should not be used in these cultures (11). This study hypothesized that microbial proteases as a safe source might be used as an alternative to trypsin. Different microbial proteases have various cleave sites, and their effects on cellular surface proteins can be similar or dissimilar to trypsin (12). These proteases are not of eukaryotic origin. The metalloproteases (MMPs) of cultured cells have various target sites in them, and their residue in cell culture is degraded by MMPs (13), which might be better tolerated by the cells.
2. Methods
2.1. Isolation of Protease Enzyme-Producing Microorganisms
The leather industry’s wastewater samples were collected, taken with a sterile container, and transported to a laboratory on ice. Then, the wastewater was diluted with phosphate-buffered saline (PBS) (ratio 1:10 to 1:1000). These dilutions were cultured on brain heart infusion (BHI) agar (Merck, Germany) and incubated at 30°C. Different colonies appeared in the media at different intervals, and the duration of the emergence of each colony in the medium was different.
2.2. Plate Assay
The proteolytic zone of proteases in substrates was measured and compared. After obtaining a single colony, the colonies were cultured in three substrate media, including Skim milk agar, gelatin agar, and casein agar (all from Merck, Germany). The plates were incubated in a 30°C incubator for 72 hours. The colonies that grow in all three substrate media were selected for further study, and the stocks were stored at -20°C (14).
2.3. Microorganism Identification
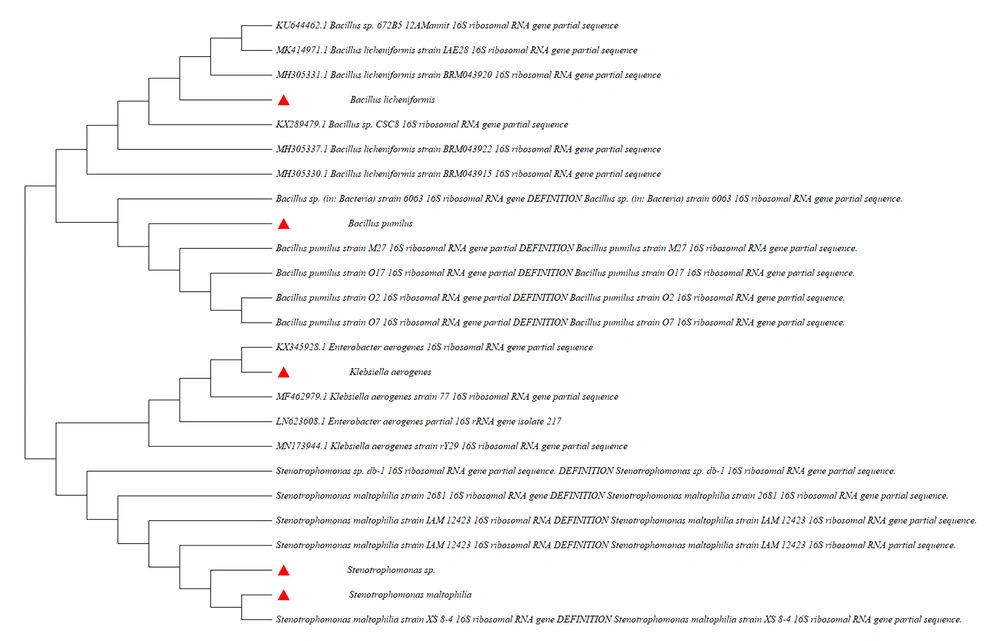
The isolated colonies were maintained in a 5 mL BHI medium and incubated at 30°C for 24 hours, and then the bacterial deoxyribonucleic acid (DNA) was extracted using the DNA Extraction Kit (Aryadaneshgene, Iran). The 16S ribosomal ribonucleic acid (rRNA) gene was amplified using the forward primer: 5΄-AGAGTTTGATCCTGGCTCAG and the reverse primer: 5΄-AAGGAGGTGATCCAGCCGCA by polymerase chain reaction (PCR). The steps of PCR included primary denaturation at 95°C for 10 minutes, 35 cycles of denaturation at 95°C for 30 seconds, annealing at 54°C for 30 seconds, extension at 72°C for 54 seconds, and final extension at 72°C for 10 minutes. Then, the PCR products were electrophoresed on the 1% agarose gel. The PCR products were sequenced and compared with other sequences in the GenBank using the National Center for Biotechnology Information (NCBI) BLAST algorithm.
2.4. Enzyme Activity Assay of the Crud Protease
The bacterial occultation cultures in 4 mL BHI broth (Merck, Germany) were incubated at 25°C for 48 hours. Then, the bacteria culture medium was centrifuged at 2000 g for 10 minutes to remove the cells from the medium. Afterward, 1 mL of the centrifuged supernatant was added to 1 mL of PBS at pH 8, and 1 mL of 2% casein (Merck, Germany) with pH 7 was added to the solution at 37°C for 10 minutes. Subsequently, 2 mL of 5 M trichloroacetic acid (TCA) (Merck, Germany) was added to stop the reaction. The casein solution that precipitated was centrifuged at 3000 g for 5 minutes. Then, 10 mL of 0.5 M sodium hydroxide (NaOH) (Merck, Germany) and 3 mL of diluted Folin reagent (Merck, Germany) (1 mL of Folin reagent diluted in 2 mL of distilled water) were added, and its optical absorbance was read at 750 nm (15).
2.5. Protease Purification
The crud protease was isolated by the filtration of bacterial culture in BHI broth, the addition of 60% ammonium sulfate, and centrifuge at 12000 g for 15 minutes. The supernatant was discarded, and the residue was further purified by a diethylaminoethyl cellulose column (15 × 300 mm) (equilibrated with 0.1 M Tris-HCl). The column was washed with the same buffer at a flow rate of 10 mL per hour, and fractions were collected (each fraction equal to 4 mL). The enzyme activity assay was carried out (15, 16).
2.6. Consequences of Different Substrates on Enzyme Activity
The effects of various substrates were measured on enzymatic activity. The multiple substrates included gelatin, casein, and keratin, which were used to determine the alteration of the isolated protease enzyme activity. The different substrates were added to 100 µL PBS. The purified enzyme (0.25 mg/100 µL PBS) was added to the substrate PBS solution and incubated at 37°C for 10 minutes. Then, 0.2 mL of 5 M TCA (Merck, Germany) was added to stop the reaction. The solution was centrifuged at 3000 g for 5 minutes, and 1 mL of 0.5 M NaOH (Merck, Germany) and 0.3 mL of diluted Folin reagent (Merck, Germany) (1 mL of Folin reagent diluted in 2 mL of distilled water) were added. The reaction optical density (OD) was read at 750 nm. The kinetic parameters were calculated using various casein concentrations as a substrate from the Michaelis-Menten equation.
2.7. Viability Determination by MTT Assay
The L929 cells were cultured in a 96-well plate (10000 cells/well) and incubated at 37°C with 5% Co2. After 24 hours, the cultured cells were detached with different extracted microbial protease enzymes (0.25 mg/100 µL normal saline) or trypsin (0.25 mg/100 µL normal saline) (Sigma, USA) as control and cultured again for 24 hours and subjected to the MTT assay. Briefly, the supernatant was discarded, and 10 µL MTT reagent (Merck, Germany) with a final concentration of 50 mg/mL dissolved in PBS was added to each well and incubated at 37°C for 2 hours. Then, the MTT reagent was removed, 50 µL of isopropanol (Merck, Germany) was added, and the OD was read at 570 nm.
2.8. Statistics
The statistical test was performed by SPSS software (version 28) and GraphPad software (version6.07). One-way analysis of variance and SPSS software (version 28) were used to determine the significant differences between the results. The significant differences between the groups were considered at less than 0.05.
3. Results
3.1. Assay of Isolated Colonies and Identification
The isolated bacteria were cultured in three substrate media. The total number of bacteria that grow and produce proteolytic zone in three media, including, Skim Milk agar, gelatin agar, and casein agar, were 24. Five isolates were selected due to their potent extracellular protease (Figure 1). The aforementioned five isolates were m1, m2, m3, m4, and m5. After specific PCR, their 16S rRNA gene was sequenced and highly similar to some data from the Nucleotide database NCBI. The m1, m2, m3, m4, and m5 isolates were Bacillus pumilus, Stenotrophomonas sp., Klebsiella aerogenes, Stenotrophomonas maltophilia, and Bacillus licheniformis, respectively.
3.2. Results of Protease Activity
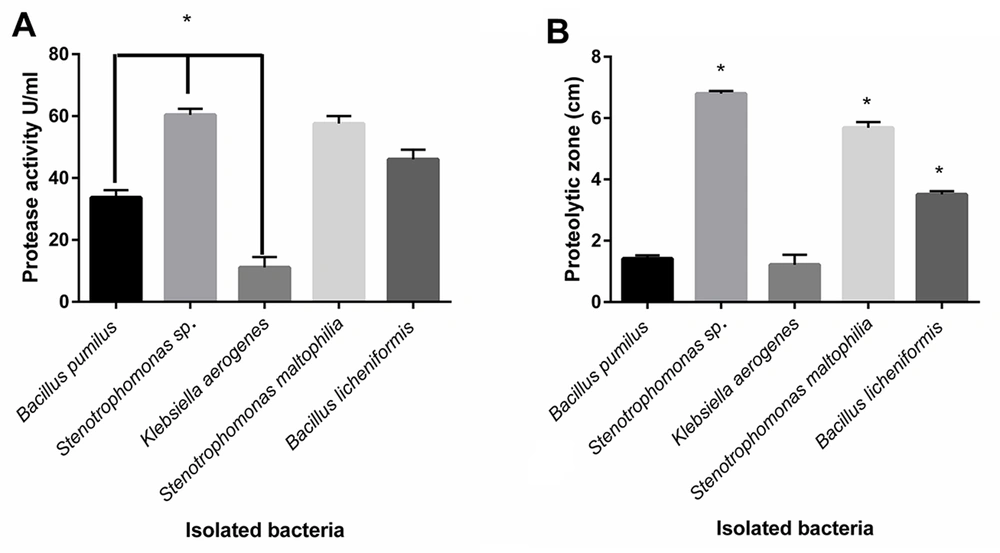
The proteolytic enzyme activities of isolated bacteria were determined by plate assay and the spectrophotometric method (14-16). The significant highest protease activity belonged to Stenotrophomonas sp., and S. maltophilia and B. licheniformis were in the following steps, respectively (P < 0.05) (Figure 2A). The protease activity values of isolated protease from B. pumilus, Stenotrophomonas sp., K. aerogenes, S. maltophilia, and B. licheniformis were 33.67, 60.34, 11.09, 57.59, and 46.04 U/mL, respectively.
Comparison of measured protease activity of isolated proteases by the spectrophotometric method; (A) Measured proteolytic zone of cultured bacteria in skim milk agar; (B) Stenotrophomonas sp., Stenotrophomonas maltophilia, and Bacillus licheniformis indicating the significant highest proteolytic zone, compared to other bacteria, respectively (P < 0.05). Stenotrophomonas sp., S. maltophilia, and B. licheniformis showed significant most excellent protease activity (P < 0.05).
The diameter zone of clearance of the Skim Milk was measured from the zone’s border to the colony’s border. The results of the measured proteolytic area indicated that all the isolated bacteria produced a proteolytic zone in plates containing Skim Milk agar, and the Stenotrophomonas sp. created the highest proteolytic zone, followed by S. maltophilia and B. licheniformis, respectively (Figure 2B).
3.3. Alteration of Enzymatic Activity Using Different Substrates
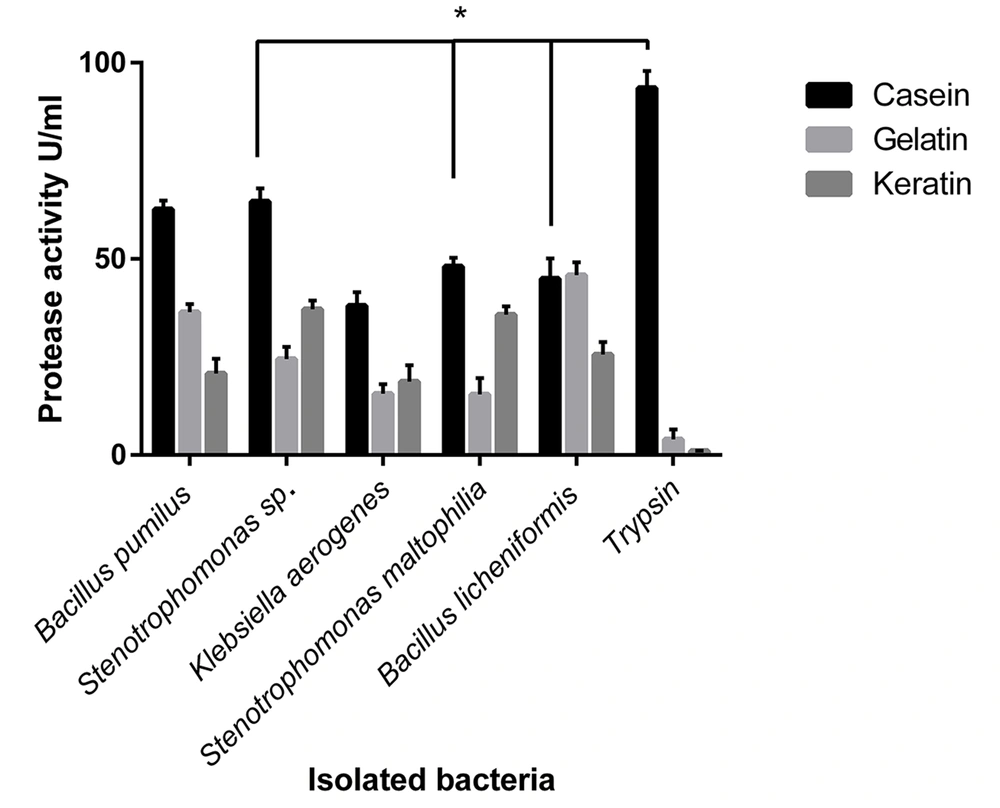
Different substrates, including gelatin, casein, and keratin, were used as protease substrates to investigate the effects of substrate on protease activity. The most protease activity among the isolated bacteria was observed for casein as a substrate, except B. licheniformis, which showed the same protease activity for gelatin (Figure 3). Bacillus pumilus, Stenotrophomonas sp., and S. maltophilia showed significant differences in protease activity against casein, gelatin, and keratin as a substrate (P < 0.05).
Effects of various substrates on the protease activity of isolated bacteria. The types of substrate affected the activity of the enzymes. The differences in observed protease activity among casein, gelatin, and keratin as substrates were significant (P < 0.05) in Bacillus pumilus, Stenotrophomonas sp., and Stenotrophomonas maltophilia. All the isolated microbial enzymes had a significantly increasing enzyme activity for casein (except Bacillus licheniformis, with similar activity for gelatin).
3.4. Effects of Substrate Concentrations on Enzyme Activity
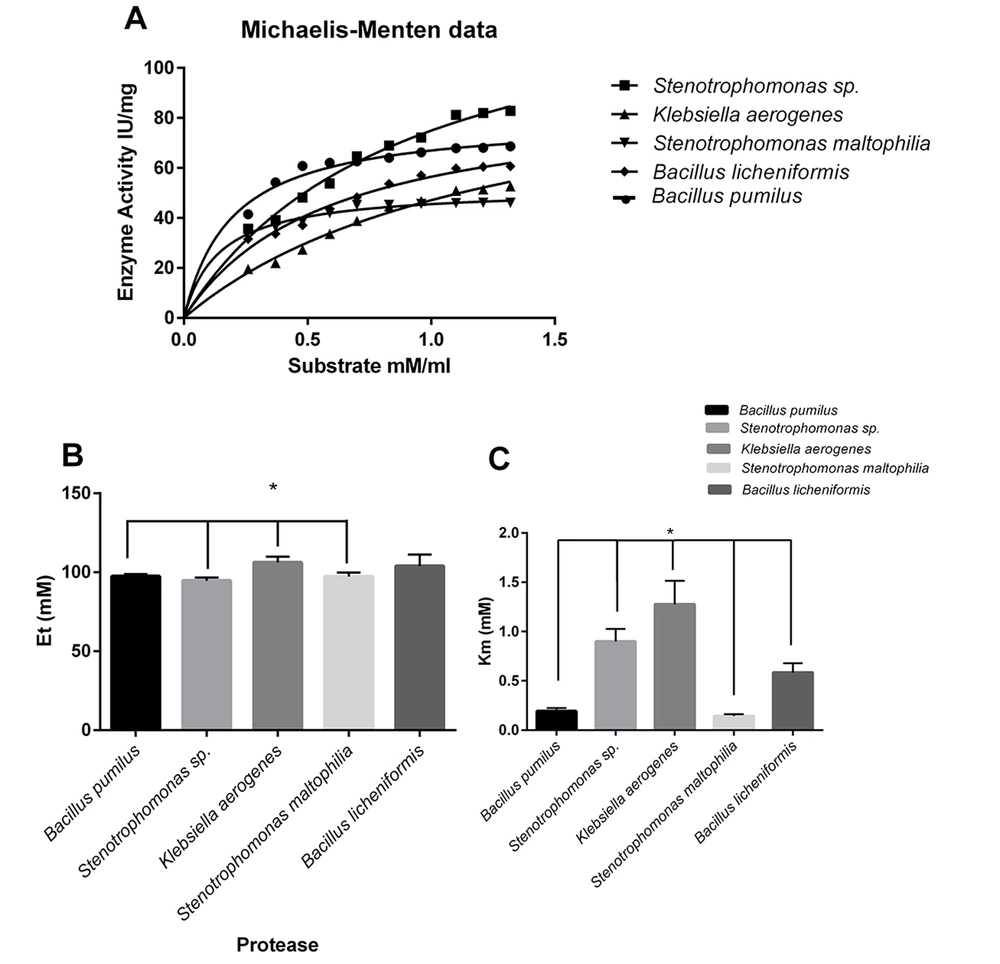
The various concentrations of casein (as a substrate) were used to determine alteration protease reaction. According to the Michaelis-Menten equation, kinetic parameters were calculated. The highest observed maximum reaction velocity (Vmax) belonged to Stenotrophomonas sp. protease, and K. aerogenes and B. licheniformis were in the following order in observed Vmax, respectively (Figure 4A).
Effect of substrate concentration on proteolytic activity; Michaelis-Menten equation microbial enzymesubstrate catalyzed reaction (A); Calculation of Et or the enzyme catalytic sites’ (mM) concentration; Casein used as a substrate to indicate the effect of substrate concentration on proteolytic activity (B); Calculation of Km for microbial proteases (the substrate concentration that saturated half of active sites of each enzyme); Km of Klebsiella aerogenes protease with a significant difference, compared to those of other proteases (C)
The Et or the enzyme catalytic sites (mM) concentration was calculated (Figure 4B).
Klebsiella aerogenes protease showed a significant difference in comparison to S. maltophilia, B. pumilus, and Stenotrophomonas sp. proteases (Figure 4B). The Michaelis-Menten constant shows the enzyme’s affinity to the substrate (Km value). The lowest Km value belonged to S. maltophilia, B. pumilus, and B. licheniformis, respectively. The lower Km content indicated the enzyme’s higher affinity to the substrate (Figure 4C).
3.5. Results of Cell Culture After Treatment by Isolated Microbial Proteases
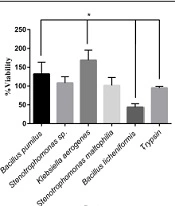
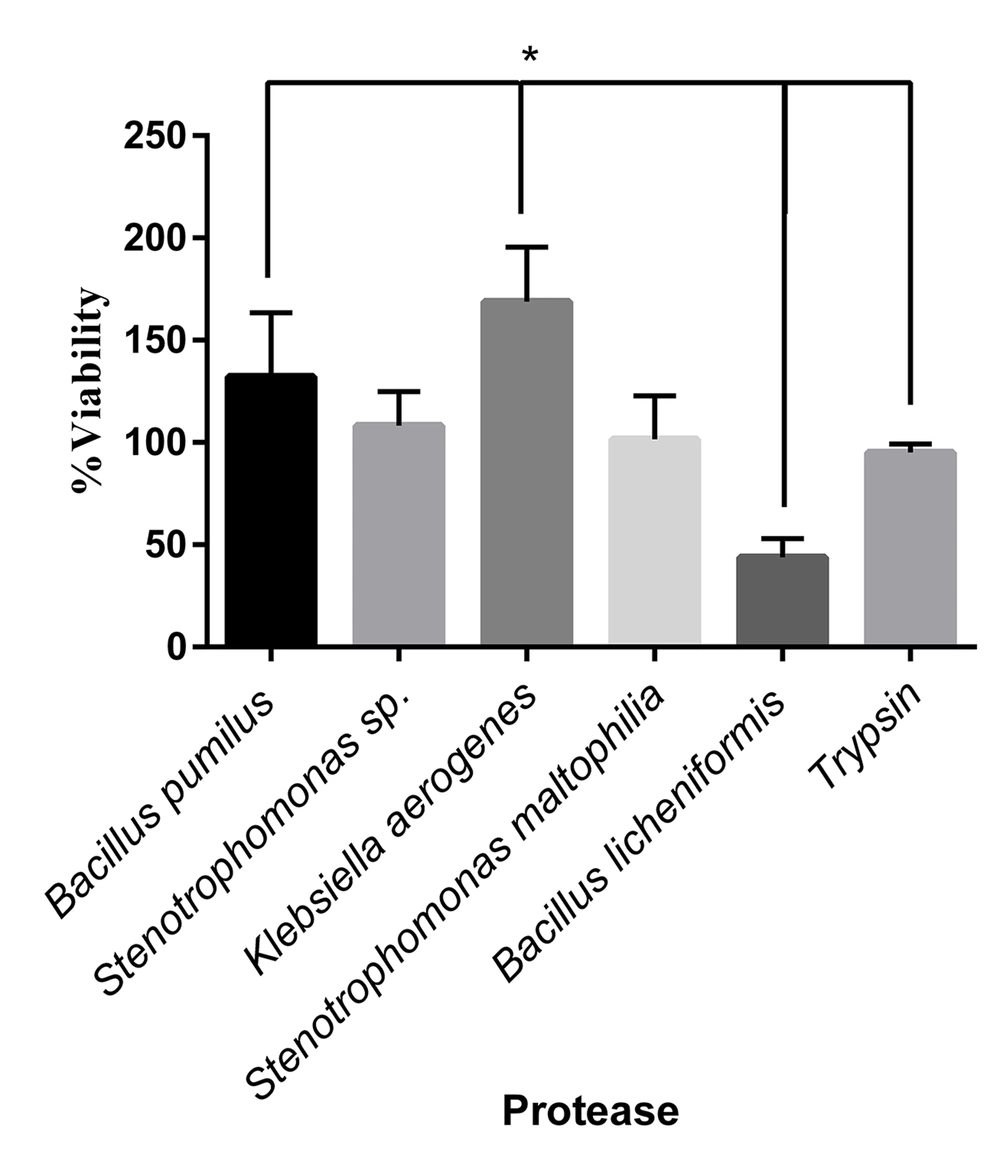
The L929 cells were cultured in a 96-well plate (10000 cells/well). After 24 hours, the cultured cells were treated with various isolated protease enzymes and trypsin as a control. The detached cells were washed with FBS and refined in a new 96-well plate. The viability of cultured cells was determined after 24 hours by the MTT assay. The L929 cells showed different responses to treated enzymes. The significant highest viability percentage was observed in cells treated with K. aerogenes protease (Figure 5). The cell viability was reduced after detachment by the extracted protease enzymes of B. licheniformis. The cells separated with Stenotrophomonas sp. and S. maltophilia proteases resulted in the same viability as trypsin.
Viability percentage in cells treated by different microbial proteases and trypsin. The cell viability increased significantly in the cells treated with Klebsiella aerogenes protease and Bacillus pumilus protease, respectively, compared to that of trypsin (P < 0.05). The Bacillus licheniformis protease decreased cell viability significantly, compared to trypsin (P < 0.05). The cell viability in treated cells by Stenotrophomonas sp. and Stenotrophomonas maltophilia were the same as trypsin.
4. Discussion
Adhesive cell culture is becoming more critical every day. The large-scale cell culture is needed for vaccine synthesis, biopharmaceutical production, tissue engineering, and increasing demand for stem cell transplantation for clinical use. All of the cell cultures should grow up gradually. In this way, cell detachment is required to transport the cells into the larger place. Trypsinization is a routine method for detaching the adherent cells in research. Trypsin manufactured from bovine or porcine pancreas should not be used in the pharmaceutical approach due to contamination by animal agents. Other methods for detaching the adherent cells are scraping (17), washing down by microjet (18), sonication (19), recombinant trypsin (20), and synthetic peptides (21). Some of the aforementioned methods reduce viability due to membrane damage, and others are not readily available or are expensive (19).
In this study, extracellular protease enzymes from five isolated bacteria were characterized. The sequencing of their 16S rRNA gene and comparing their sequences to the data in the Nucleotide database of NCBI showed that they were Bacillus sp., Stenotrophomonas sp., K. aerogenes, S. maltophilia, and B. licheniformis. The proteases of the isolated bacteria were extracted, and the L929 cell line, routinely used for cytotoxicity study, was exposed to microbial proteases and trypsin in parallel. The detached cells were cultured again, and their viability was evaluated. The treatment of the L929 cells with the Stenotrophomonas sp. protease and S. maltophilia protease resulted in the same as trypsin. It was observed that the treatment of the L929 cells with the B. licheniformis protease reduced cell viability. While treating the cells with K. aerogenes and B. pumilus, protease enzymes enhanced cell viability after treatment, compared to trypsin. This finding should be due to differences in microbial protease activity. The isolated proteases in this study showed significant differences in protease activity.
The isolated protease from B. pumilus had 33.67 U/mL activity in this study. However, in previous studies on B. pumilus, the protease activity was 35.437 U/mL (22) to a maximum of 2000 U/mg (23), which was significantly higher than the present study. The Stenotrophomonas sp. protease determined at 60.34 U/mL in this study was previously reported as 40 mg/mL in the isolate in 2017 (24) to 157.50 U/mg (25). In this study, the K. aerogenes protease was 33.27 U/mL, previously detected at 14.24 U/mL (26). Similar differences were observed in S. maltophilia protease activity which was 57.59 U/mL in this study; nevertheless, it was previously determined at 1.87 U/mg (27) to 418 U/mL (28). The protease detected from B. licheniformis had 46.04 U/mL protease activity in this study which was previously reported from 3.2 U/mL (29) to 5100 U/mL (30).
In addition, each protease showed a preferred substrate to cleave, and there were significant differences between isolated enzymes’ protease activity concerning the used substrates. For example, B. pumilus had a high protease activity in casein, and its protease activity was significantly reduced in gelatin. It showed the lowest proteolytic activity in keratin used as a protease substrate. Although Stenotrophomonas sp., S. maltophilia, and K. aerogenes similarly showed the highest protease activity for casein, the preferred substrates were keratin in the subsequent step and gelatin, respectively. Bacillus licheniformis had an equal protease activity in casein and gelatin substrates and a significantly reduced protease activity observed in keratin. Trypsin had a quite different activity pattern. It showed increased protease activity for casein, a minimum activity for gelatin, and almost no protease activity for keratin.
Comparing cellular viability outcomes after treatment by proteases and their effects on substrates makes it possible to select a more suitable protease for use in cell cultures. This study showed that the protease activity for gelatin as a substrate could lead to choosing the better enzyme for trypsin replacement. The enzymes, which had lower protease activity (< 30 U/mL) for gelatin as a substrate, enhanced L929 cell viability after the treatment. These outcomes can be used as an indicator to find the better applicable protease enzymes instead of trypsin for cell detachment, which is an obligatory step for transferring cells in cell culture experiments.
Another critical point is the optimization of microbial enzyme concentrations. In this study, the concentration of all used isolated microbial proteases was the same as trypsin. Optimization according to the protease activity of microbial proteases resulted in minimizing damage to the cell membrane and extracellular matrix of cells during displacement and led to enhanced cell viability after separation and reduced unwanted side effects. The microbial enzymes have the most crucial advantage. After cell detachment with microbial enzymes, rinsing the cells with PBS is sufficient to remove their residue and does not require the addition of FBS (31).
4.1. Conclusions
The medical cell cultures need animal origin-free cultures, and microbial enzymes are the best, safe, and available alternative for trypsin replacement in cell cultures. The proteolytic activity of microbial enzymes in different substrates differs from trypsin; however, their function in the detachment of the cells is similar to trypsin (or better) in cell cultures. In addition, they can be helpful in cell lines that are hard to detach by trypsin. For the removal of their residue, rinsing the cell is sufficient.