1. Background
Acinetobacter baumannii, as a nosocomial pathogen, causes various infections in different anatomical sites of the human body, such as skin, bloodstream, and urinary tract, with high mortality and morbidity rates (1, 2). This bacterium has a high incidence in immunocompromised individuals, especially those who have had a prolonged (≥ 90 days) hospital stay (3). Although this Gram-negative coccobacillus was considered a low-grade pathogen in the past, it has become a major public health concern nowadays due to its high resistance to most clinically available antibiotics. In recent years, various strategies, including vaccination (4), monoclonal antibodies (5), phage therapy (6), iron chelators, and gallium nitrate (7), have been developed to control infections with this bacterium. Of all the aforementioned strategies, vaccination might be the most effective approach to prevent infections associated with this bacterium, especially in target groups, such as the elderly, soldiers, and immunocompromised patients (8).
Multicomponent antigens and purified recombinant proteins are the two main vaccine strategies against A. baumannii. Although these immunogenic candidates showed acceptable results in animal models, they are not yet satisfactory for human injection (9, 10). Less attention has been paid to the extracellular proteins of A. baumannii as potential immunogenic candidates. According to the results from different bioinformatics databases and studies, this group of proteins plays an important role in the pathogenesis of A. baumannii (11).
With the advances in bioinformatics and the presence of the whole genome sequence of bacteria in various bioinformatics databases, such as Vaxign, the ideal promising immunogenic candidates can be found by reverse vaccinology method and in silico approach. In this context, it is worth mentioning that Meningococci group B represents the first example of the successful application of reverse vaccinology (12). With the consideration of all the above-mentioned information, this study investigated a dataset of A. baumannii whole-genomes to analyze extracellular or secreted proteins as putative immunogenic candidates.
2. Methods
2.1. Identification and Selection of Putative Immunogenic Candidates
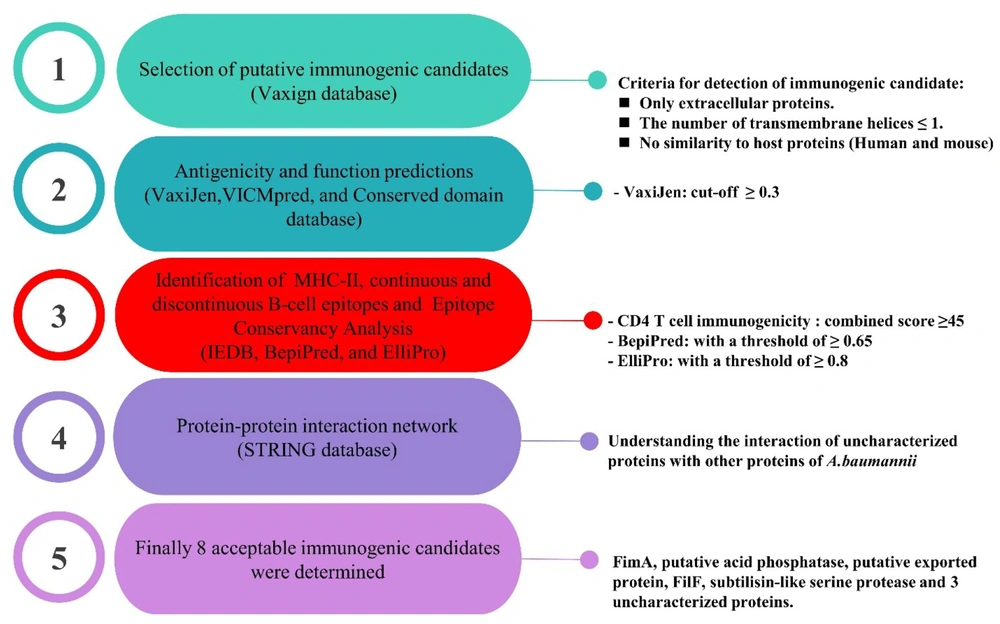
In the first step, Vaxign (vaccine design webserver), a vaccine target prediction and analysis system based on the principle of reverse vaccinology, was used to select ideal putative immunogenic candidates. A total of 35 genome sequences of A. baumannii strains were selected from this database. Subsequently, the A. baumannii strain ACICU (belonging to ST2Pas) was determined as a reference strain. In the next step, the analysis was restricted to only extracellular proteins present in all genomes. In addition, other criteria, such as the number of transmembrane helices ≤ 1 and no similarity to human and mouse proteins, were considered (Figure 1) (13).
2.2. Antigenicity Measurement and Function Prediction of Putative Immunogenic Candidates
The VaxiJen database is a helpful tool to predict protective antigens and subunit vaccines. This database was used to measure the antigenicity of putative immunogenic candidates with a cut-off ≥ 0.3 (14). The VICMpred is a useful database for the prediction of functional classes (e.g., virulence factors, cellular processes, and metabolism molecules). The amino acid sequence of each putative immunogenic candidate was submitted to this database to predict their functions and scores (15).
2.3. Identification of Major Histocompatibility Complex Class II Binding Sites
The prediction of CD4+ T cell immunogenicity was performed using the Immune Epitope Database (IEDB) Analysis Resource. In this study, the maximum combined score threshold was 50%, and a combined score ≥ 45 was acceptable. In the next step, the number of major histocompatibility complex class II (MHC-II) epitope binding sites was divided by the number of amino acids for each protein and expressed as ratios (16, 17).
2.4. Identification of Continuous and Discontinuous B-Cell Epitopes
BepiPred predicts the location of continuous B-cell epitopes from amino acid sequence using a Hidden Markov Model. For this purpose, the sequence of each protein in FASTA format with a threshold of ≥ 0.65 was submitted (18). In this part, similar to the MHC- II epitopes, the number of continuous B-cell epitopes was divided into the number of amino acids for each immunogenic candidate and reported as ratios.
ElliPro is another database for antibody epitope prediction. This database was used to predict the B-cell discontinuous epitopes of each putative immunogenic candidate with a threshold of ≥ 0.8 (19). The three-dimensional (3D) structure prediction files were obtained using Robetta servers and refined using the ModRefiner server. In the next step, their quality was checked using QMEAN Swiss Model. The Protein Data Bank (PDB) files of the putative immunogenic candidates were shown in the Jmol software (version 14) for illustration. This software is an open-source Java viewer for chemical structures in three dimensions (20). In the next step, the B-cell discontinuous epitopes of each immunogenic candidate were displayed on the surface of the proteins with different colors.
2.5. Evaluation of MHC-II and B-Cell Epitopes Conservancy among Different Strains of Acinetobacter baumannii
In this part, the MHC-II and B-cell epitopes were transmitted to Epitope Conservancy Analysis in IEDB Analysis Resource, and their conservation among different strains of A. baumannii was analyzed (21).
2.6. Identification of Conserved Domains in Selected Proteins
Conserved Domain Database is a part of NCBI’s Entrez query that provides an annotation of protein sequences with the location of the conserved domain. This database was used to predict the conserved domain of each putative immunogenic candidate (22).
2.7. Protein-Protein Interaction Network
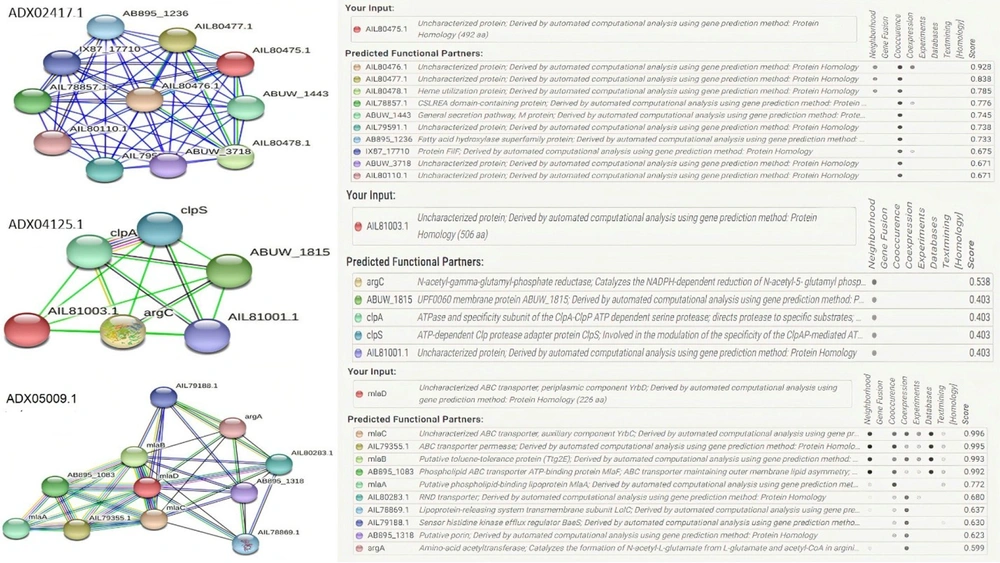
The STRING is a helpful database for understanding how proteins interact with each other. This database contains information from numerous sources, including experimental data, computational prediction methods, and public texts. In this part, the STRING database was used to understand the interaction of uncharacterized proteins with other A. baumannii proteins. This approach can be effective for the function estimation of unknown proteins (23). The red spheres show the target proteins, and others represent other proteins in relation to the hypothetical proteins. Each interaction has a specific color (light blue: from curated databases, violet: experimentally determined, green: gene neighborhood, red: gene fusions, blue: gene co-occurrence, yellow: text mining, black: coexpression, grey: protein homology).
3. Results
3.1. Identification and Characterization of Putative Immunogenic Candidates
In this study, 19 extracellular proteins were discovered in the Vaxign database. The accession numbers and other characteristics of all 19 extracellular proteins are shown in Appendix 1 (see Supplementary File). All of these proteins have a ≤ 1 transmembrane helix. The proteins that have similarities to host proteins were excluded. The next step was to measure the antigenicity of the remaining proteins, and the proteins with the highest antigenicity value were considered potential immunogenic candidates. These proteins were FilF (ADX02311.1), FimA (ADX03638.1), putative acid phosphatase (ADX03936.1), putative exported protein (ADX04185.1), subtilisin-like serine protease (ADX04983.1), and three uncharacterized proteins (i.e., ADX02417.1, ADX04125.1, and ADX05009.1). According to VICMpred prediction, the aforementioned proteins are categorized as virulence factors, metabolic molecules, and cellular processes. Table 1 shows the complete results of this investigation.
| No. | Protein Name | Protein Domain | Subcellular Localization (PSORTb) | MHC-II 15-mer Epitopes Sites (CS ≥ 45) | T-Cell Epitope Ratio | B-cell Epitopes (T = 0.65) | B-Cell Epitope Ratio | VaxiJen Score | VICMpred |
|---|---|---|---|---|---|---|---|---|---|
| 1 | FilF | No domain | Extracellular | 20 | 0.030 | 10 aa | 0.0154 | 0.64 | Virulence factors (2.18) |
| 2 | Putative uncharacterized protein | No domain | Extracellular | 7 | 0.014 | 18 aa | 0.036 | 0.72 | Virulence factors (2.6) |
| 3 | FimA | Fimbrial superfamily protein | Extracellular | 9 | 0.026 | 6 aa | 0.017 | 0.66 | Virulence factors (0.86) |
| 4 | Putative acid phosphatase | Polyphosphatase and 5'/3'-nucleotidase SurE | Extracellular | 7 | 0.021 | 20 aa | 0.061 | 0.62 | Metabolism molecule (2.57) |
| 5 | Putative uncharacterized protein | No domain | Extracellular | 14 | 0.027 | 8 aa | 0.015 | 0.54 | Virulence factors (0.32) |
| 6 | Putative exported protein | TolA-binding protein | Extracellular | 8 | 0.027 | 37 aa | 0.12 | 0.68 | Virulence factors (1.50) |
| 7 | Uncharacterized protein | MlaD protein superfamily | Extracellular | 7 | 0.030 | 30 aa | 0.13 | 0.73 | Cellular process (1.54) |
| 8 | Subtilisin-like serine protease | Peptidase S8 family | Extracellular | 11 | 0.025 | 3 aa | 0.007 | 0.57 | Metabolism molecule (2.17) |
Abbreviation: MHC-II, major histocompatibility complex class II.
3.2. Identification of MHC-II and Continuous and Discontinuous B-Cell Epitopes of Putative Immunogenic Candidates
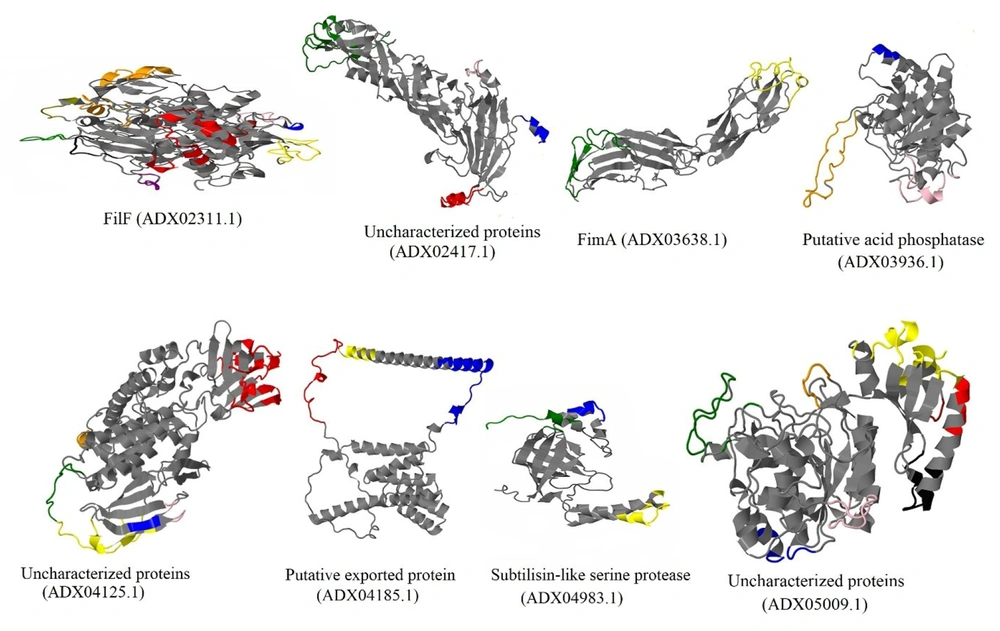
Table 1 shows a list of the ratios of MHC-II and linear B-cell epitopes of each immunogenic candidate. In the case of discontinuous B-cell epitopes, the predicted epitopes were plotted on a 3D structure of the putative immunogenic candidates in Figure 2 (the full results of discontinuous B-cell epitopes prediction were listed in Appendix 2).
Predicted discontinuous B-cell epitopes of putative immunogenic candidates against Acinetobacter baumannii shown on a three-dimensional structure of proteins; FilF, uncharacterized protein (ADX02417.1), FimA, putative acid phosphatase, uncharacterized protein (ADX04125.1), putative exported protein, subtilisin-like serine protease, and uncharacterized proteins (ADX05009.1) with 10, 4, 2, 3, 7, 3, 3, and eight conformational epitopes, respectively.
3.3. Evaluation of MHC-II and B-Cell Epitopes Conservancy among Different Strains of Acinetobacter baumannii
Table 2 shows a summary of the results of MHC-II and B-cell epitopes conservancy among different strains of A. baumannii with the percentage of conservation. FimA, putative exported protein, and uncharacterized proteins, including ADX04125.1 and ADX05009.1, had high conserved epitopes.
| Protein Name | MHC-II Epitopes | Conservation (%) | B-cell Linear Epitopes | Conservation (%) |
|---|---|---|---|---|
| FilF | YLERNVFKTSGNVKT | 58.93 | CTPTTSDNGAEDS | 64.29 |
| REIQSTTPLRLSFSN | 73.21 | TAVNGKSKEINP | 67.86 | |
| YLLTTKVKPAKLNVN | 69.64 | KGSAIVIDNKVQN | 67.86 | |
| DKNDETIKVAIALIK | 92.86 | TDIKPNSTTTDMS | 75.00 | |
| YKLLVQTTNLSNAGV | 69.64 | AATFNDPLNGDQS | 58.93 | |
| IVIDNKVQNPYLLTT | 67.86 | |||
| FLGNVNLDTIGKFTA | 92.86 | |||
| FimA | MKKMLIKSGLLITSF | 90.00 | LNYPVSSIGNNV | 98.00 |
| LITSFFIYNQTYANC | 100.00 | DSTDFSGYYPYKRSLTPNTTYTLS | 69.00 | |
| TTYTLSPGYFVMEVI | 97.00 | NGGENNSGVPTSNT | 97.00 | |
| SPGYFVMEVIKTAAT | 97.00 | ISDTSNSQVINN | 72.00 | |
| TTTVLSSSPILIASS | 97.00 | |||
| VELLWNMNGANNPIR | 92.00 | |||
| PLTARYYQTASNVTA | 94.00 | |||
| Putative acid phosphatase | ALNILLVNDDGLTSN | 100.00 | NDKQIAAENGCHNGAAPIGAPAAGTFTKAGY | 86.00 |
| GRGGAIVMYSSTTIT | 95.00 | NTVDDKTLNNPNS | 84.62 | |
| IVMYSSTTITADNDK | 95.00 | |||
| PKNLTTSTPFAFSKI | 98.00 | |||
| YKLKFQVTKDNKGNN | 73.00 | |||
| GQEWLKIRLKSLFNK | 84.00 | |||
| Putative exported protein | HSLLFIALMSTTSLY | 98.00 | PDSATQDDSSNATSDNTTPASAPAPQTTESNKVAAVPATQTSEQQPSAPTT | 37.00 |
| IALMSTTSLYANIPI | 99.00 | AQNQSNSLEL | 71.00 | |
| TTSLYANIPIESRGL | 99.00 | |||
| IAPMQNFIKNHPNSI | 93.00 | |||
| NFIKNHPNSIYTGNA | 96.00 | |||
| Subtilisin-like serine protease | GIFVIIFGIALFFLA | 91.00 | TQATPAQQKTMEQQLISNMNSITSIDE | 30.00 |
| LFFLAMKVSGLVGTN | 96.00 | GKVAAGSSSAEEKAPASTDSS | 41.00 | |
| GRVDSITLDPVTRLA | 97.00 | |||
| MEQQLISNMNSITSI | 34.00 | |||
| DEDAYIMVATNGLLG | 99.00 | |||
| EKYLKIVPGGGLNYL | 50.00 | |||
| Uncharacterized protein (ADX02417.1) | MRQFTSLQVAILALG | 55 | TDSANLEKLRDNSSNI | 87.00 |
| GQALMSMSYIAPTDS | 93.00 | DANGNPLPMSNEDRASS | 58.00 | |
| EAELEINTNIRKLQL | 87.00 | GTENTTTPNGIN | 61.00 | |
| VVGLRLSAEKFIGLL | 74.00 | NIYVGHSSSYPVDGTVQYNAAGPHDPAYPEPTGIYTQGGK | 78.00 | |
| ISGYMKVQSDSSGLI | 80.00 | |||
| TANLNLTQSLGLIH | 97.00 | |||
| NLTQSLGLIHNLPIN | 98.00 | |||
| LGLIHNLPINSPMYL | 100.00 | |||
| SPMYLALQNQMLQWP | 93.00 | |||
| LFPQISAAVSAYLQA | 51.00 | |||
| Uncharacterized protein (ADX04125.1) | ASQDVYRSLLNAFKD | 91.00 | SSGGSSNTPKPSEPT | 100.00 |
| YRSLLNAFKDTNIPS | 99.00 | ADLQDQSHLDPIG | 100.00 | |
| TLLDIAANQKVTRDY | 93.00 | GDYRRAVG | 100.00 | |
| GNTSEFFRIDGAGDY | 97.00 | NSSSSEPNNSTQQYN | 100.00 | |
| FFRIDGAGDYRRAVG | 99.00 | |||
| CQLNFRANGQIELIK | 98.00 | |||
| IELIKGSQSYRSALS | 92.00 | |||
| QQYNFIQLHIKANKI | 94.00 | |||
| IQLHIKANKIVSAQA | 94.00 | |||
| Uncharacterized protein (ADX05009.1) | LERRSYIATNKKNLQ | 99.00 | VVYSFDSAPDD | 100.00 |
| AGGVQVLRDYNNLPL | 97.00 | VKYTSECSDS | 99.00 | |
| VLRDYNNLPLAFYRI | 99.00 | IGGVSWGNPVKCSDPTTAADKVACFSNG | 97.00 | |
| NDPNVKAVYPNRINR | 98.00 | |||
| TKTKIIGIDVFRKVR | 96.00 | |||
| IGIDVFRKVRSQGKW | 97.00 | |||
| SSYGTAFANARAAGV | 98.00 | |||
| IALLRANSVSPTESI | 97.00 | |||
Abbreviation: MHC-II, major histocompatibility complex class II.
3.4. Protein-Protein Interaction Network
According to the results from the STRING database, the uncharacterized protein with accession number ADX02417.1 had co-occurrence and neighborhood interactions with heme utilization protein. Another uncharacterized protein with accession number ADX04125.1 had neighborhood interactions with ClpA, ClpP, and ClpS. The CLP protease family is a family of serine peptidases. This class of enzymes cleaves peptide bonds in proteins, with serine serving as the nucleophilic amino acid at the (enzyme) active site (24). The last uncharacterized protein, ADX05009, belongs to the MlaD protein superfamily. This family of proteins contains MlaD, which is a component of the Mla pathway, an ATP-binding cassette transport system that maintains the asymmetry of the outer membrane (OM) (Figure 3) (25).
4. Discussion
Acinetobacter baumannii is an emerging opportunistic pathogen responsible for healthcare-associated infections and has become a global problem (26). Due to the frequent outbreaks caused by multidrug-resistant A. baumannii, the development of a safe and effective vaccine appears necessary. Despite all efforts to date, there is still no licensed vaccine, and studies continue to find an ideal vaccine candidate (27). In terms of vaccinology, an ideal or promising immunogenic candidate should be exposed on the bacterial surface (OM or extracellular proteins) for host immune system induction, have a high antigenicity value, be conserved across circulating strains, and be expressed during bacterial infection. In addition, the ideal immunogenic candidates should have an important role in bacterial pathogenesis (28). In the case of A. baumannii, most studies focused on outer membrane proteins (OMPs), such as OmpA, OmpW, and Omp22 (29), and extracellular proteins were underestimated. However, studies showed that the consideration of OMPs as immunogenic candidates had some obstacles and limitations. For example, OmpA is the most abundant OMP in A. baumannii and is involved in host-cell recognition, biofilm formation, and antibiotic resistance. Nevertheless, the coexistence of different OmpA variants and subvariants in the A. baumannii population can severely limit the efficacy of a vaccine (30).
However, the present study focused on the extracellular proteins of A. baumannii and selected the best of them according to the defined criteria, such as transmembrane helix, antigenicity score, and high adhesion probability. The current study showed that extracellular proteins, such as FilF and FimA, can be considered putative immunogenic candidates. In line with the results of the present study, Singh et al. presented FilF protein as a putative vaccine candidate in an in vivo experiment. The aforementioned study showed that immunization with FilF can produce a high antibody titer and provide 50% protection against a standard lethal dose of A. baumannii (31). The expression of FimA, a type 1 fimbrial protein, increases during the bacterial infection process and leads to the association of bacteria to biofilm formation. In this context, the efficacy of FimA and FimH for vaccine development was previously investigated (32).
In line with the results of the current study, an in silico study conducted by Zadeh Hosseingholi et al. showed that proteins involved in pilus assembly and four hypothetical proteins were likely immunogenic candidates. The aforementioned results underscored the importance of the pilus assembly system in the pathogenesis and immunogenicity of A. baumannii. Furthermore, the aforementioned data suggest that in silico approaches might be helpful in discovering likely drug and immunogenic candidates that have been ignored up to now (33).
Moreover, putative acid phosphatase, putative exported protein, and subtilisin-like serine protease were determined as desirable immunogenic candidates. The acid phosphatase enzyme is able to dephosphorylate various organic substances (34). This enzyme plays a fundamental role in Gram-negative bacteria (35). The putative acid phosphatase has a SurE domain which is expressed when bacterial cells are under environmental stress and in the steady growth phase of bacteria (36). The putative exported protein has a TolA-binding domain which has a vital role in the import mechanisms for the uptake of bacteriocins and the deoxyribonucleic acid of filamentous bacteriophage (37). The subtilisin-like serine proteases are widespread enzymes among bacteria. For example, Mycosin-1, a subtilisin-like serine protease in Mycobacterium tuberculosis, is expressed after macrophages infection and is submitted to proteolytic processing (38).
This study identified three uncharacterized proteins as potential immunogenic candidates according to the defined criteria. The obtained results from the STRING database demonstrated that these proteins had a role in nutrition (heme utilization), peptide bond cleavage (serine peptidases), and cellular processes (MlaD protein), respectively. Another in silico approach by Mobarak Qamsari et al. indicated that the ZnuD protein is a potentially suitable antigen for subunit vaccine development. This protein belongs to TonB-dependent receptors and is involved in zinc acquisition (39). Among the aforementioned proteins, the uncharacterized protein with the MlaD protein superfamily domain is more desirable for further investigation due to its high score of MHC-II and B-cell epitopes ratios. MlaD stands for the maintenance of OM lipid asymmetry. In Gram-negative bacteria, the OM has an asymmetric structure, with lipopolysaccharides on the outer surface and phospholipids on the inner surface. This unique feature makes Gram-negative bacteria resistant to numerous toxic chemicals and antibiotics (40).
In the present study, in addition to the immunogenic candidates, several MHC-II and B-cell epitopes were identified. These epitopes were surface exposed and might robustly induce the host immune system. Several studies have been conducted on the antigenic epitopes of A. baumannii. Girija, for example, targeted the GacS protein involved in citrate utilization in A. baumannii and proposed five antigenic peptides as promiscuous vaccine candidates. It should be considered that these epitopes are valuable resources for multiepitope target design (41).
4.1. Conclusions
It appears that OMPs are suitable immunogenic candidates. However, due to some disadvantages of these proteins (e.g., antigenic variation), the consideration of extracellular proteins might compensate for the disadvantages and strengthen the vaccine formulation. Furthermore, this study proposed a list of potent immunogenic candidates of extracellular proteins. In addition to finding putative immunogenic candidates, this in silico approach provided new insights into the discovery of new virulence factors of A. baumannii that were previously undiscovered or ignored. However, discovering the effective immunogenic candidates or virulence factors requires further studies in animal models and role confirmation assays, such as site-directed mutagenesis.
4.2. Limitations
The results obtained from this study were only based on bioinformatics investigation; therefore, in vivo and in vitro investigations are needed to confirm the obtained results.