1. Background
Infectious diseases induced by viruses and bacteria have been a major challenge to the health systems worldwide; hence, the generation of drug-resistant viral strains to the present antibacterial and antiviral drugs has posed an essential need for discovering and developing new effective inhibitors for viral and bacterial pathogens (1, 2).
Human immunodeficiency virus type 1 (HIV-1) has challenged the health and global economies worldwide (3). The World Health Organization (WHO) estimates that above 38 million people are infected with HIV, and 33 million infected patients would die even after the availability of the potential drugs in the market (4). However, there is a demand for new drugs with a new structure and a mechanism of action. In general, four main categories of drugs can be used in the HIV treatment, including protease inhibitors (PIs), nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), and integrase strand transfer inhibitors (INSTIs) (5). Although many therapeutics have been developed for the HIV treatment, discovering new types is of great importance due to the emergence of drug-resistant HIV-1 mutant strains, toxicity, and costs.
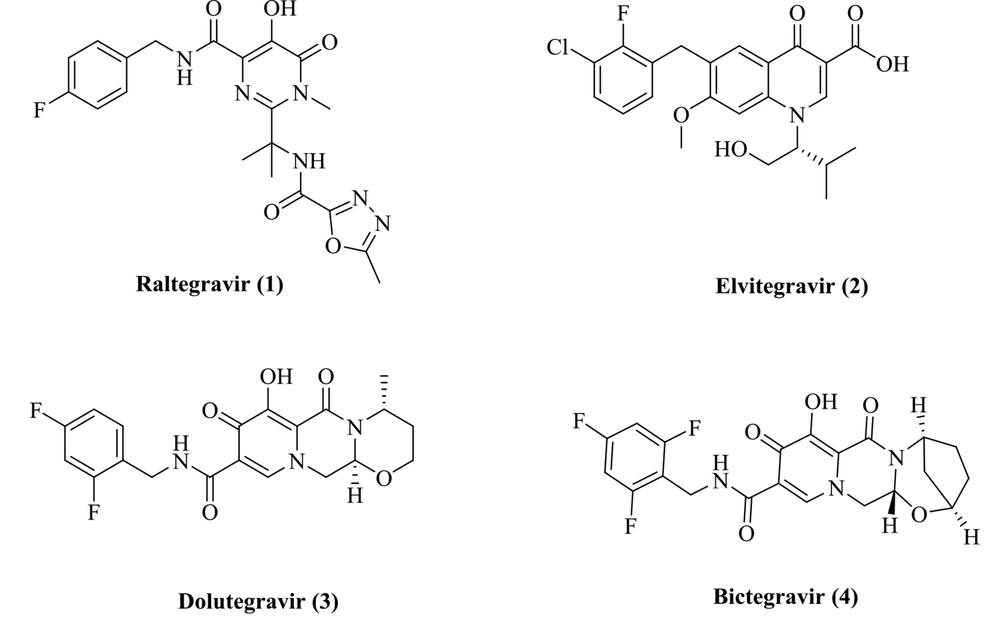
The HIV-1 integrase plays a critical role in the HIV-1 replication, and its function is to catalyze the integration process. Integration operation includes two steps entitled 3'-processing and strand transfer (ST) (6). First, a dinucleotide from each 3'-end of the viral cDNA is removed by IN (3'-processing), and the 3’-ends of the viral DNA are then integrated into the human DNA (strand transfer) (7). The integration reaction fully depends on two Mg2+ ions in the IN active site and interacts with three acidic amino acids (Asp64/Asp116/Glu152). Accordingly, the chelation of the Mg2+ cofactors can prevent enzyme ligation function (8). Many INSTIs with the metal chelation potential have been developed and reported (9, 10). Relevant research has resulted into the identification of four FDA-approved INSTIs inhibitors: Raltegravir, elvitegravir, dolutegravir, and bictegravir (Figure 1) (11-14). All INSTIs possess a planar chelating moiety interacting with Mg2+ ions and an aromatic group orienting into a hydrophobic pocket (15).
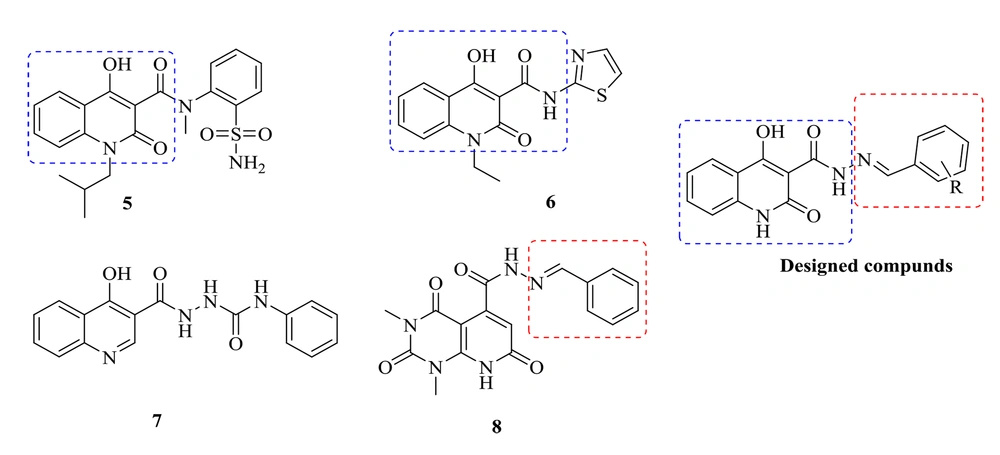
medicinal chemists and pharmacologists have recently addressed quinolines and their various substituted functionalities in many studies. Quinoline derivatives possess a variety of biological properties, including anti-HIV, antibacterial, antiviral, anti-inflammatory, anticancer, antihypertensive, analgesic, and miscellaneous properties (16). The 4-hydroxy-2-oxo-1,2-dihydroquinoline scaffold is one of the quinoline derivatives with HIV IN inhibitory and antibacterial activities, exemplified by compounds 5 and 6, respectively. Sechi et al. reported that Compound 5 inhibited both strand transfer activities and the 3′-processing of IN with IC50 = 16 ± 6 and IC50 = 40 ± 3 μM, respectively (Figure 2) (17). Compound 6 showed promising antibacterial activities (18). Accordingly, a 4-hydroxy-2-oxo-1,2-dihydroquinoline scaffold was selected to design new anti-HIV-1 and antibacterial compounds. Previously, we developed some 4-hydroxyquinoline and pyridopyrimidine derivatives containing a carbohydrazide-type framework which led to promising anti-HIV agents (19, 20). In this research, new compounds were designed by changing the amide of a 4-hydroxy-2-oxo-1,2-dihydroquinoline core with the bioisoster carbohydrazide. Furthermore, an arylidene fragment was attached to the central core to improve anti-HIV-1 activities. Accordingly, a series of N'-arylidene-4-hydroxy-2-oxo-1,2-dihydroquinoline-3-carbohydrazide derivatives were designed, synthesized, and tested for the cell-based anti-HIV-1 replication assay. The compounds were also examined for their in vitro antibacterial potencies against the several bacterial strains inducing opportunistic infections in HIV patients. Further, a docking study was conducted to analyze how the newly synthesized chemicals interact with the catalytic domain of HIV-1 IN.
2. Methods
2.1. General
All chemicals and solvents in this project were purchased from Merck AG and Aldrich Chemical. Thomas-Hoover capillary apparatus was used to determine melting points. Infrared spectra were obtained using a Perkin Elmer Model 1420 spectrometer, and 1H-NMR spectra were acquired by a Bruker FT-500 MHz instrument (Brucker Biosciences, USA) with TMS as the internal standard. Chloroform-D and DMSO-D6 were used as solvents. Coupling constant (J) values were measured in hertz (Hz), and spin multiples are presented as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), and br (broad). A 6410Agilent LCMS triple quadrupole mass spectrometer (LCMS) with an electrospray ionization (ESI) interface was used to perform mass spectral measurements, and there was a Costech 4010 elemental analyzer to perform the C, H, and N elemental analyses. The microanalysis values of C and H were within ± 0.4% of the theoretical values.
2.2. Synthesis of Ethyl 4-Hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carboxylate (10)
Isatoic anhydride (9) (10 g, 61.5 mmol) and diethyl malonate (49 mL, 300 mmol) were reacted in dry DMF (100 mL) and warmed at 85°C for 5 hours. TLC (thin-layer chromatography) was used to monitor the reaction completion. When the reaction was completed, the mixture was cooled. The reaction mixture was added to a mixture of ice and water; the obtained precipitate was filtered and washed with water: Yield: 40%, pale brown powder, mp: 134°C; IR (KBr): 1750, 1730 (C=O), 2700-3200 (OH) cm-1; LC-MS (ESI): m/z 234 [M+H]+.
2.3. Synthesis of 4-Hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (11)
Compound 10 (20 g, 55.7 mmol) was suspended in ethanol (30 mL), to which hydrazine hydrate (10 mL, 33 mmol) was added and stirred under reflux for 2 hours. When the reaction was completed, the white suspension was filtered. The precipitate was washed with ethanol and dried under a vacuum: Yield: 90%, white powder, mp: 152°C; IR (KBr): 1750, 1740 (C=O), 2800 (NH) cm-1; LC-MS (ESI): m/z 220 [M+H]+.
2.4. General Procedure for the Synthesis of Compound 12a-o
A solution of Compound 11 (1 mmol) in absolute ethanol (5 mL) was prepared, and one drop of 98% H2SO4 was then added to the solution. After that, the reaction continued by adding benzaldehyde derivatives (1.1 mmol) to the mixture and refluxed for 2 hours. When the reaction was completed (monitored with TLC), the reaction temperature was lowered in an ice bath, and the obtained precipitates were filtered. After washing with cold ethanol, the precipitate was crystallized in absolute ethanol (average yield: 90%).
2.5. Benzylidene- 4-hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12a)
mp: 225°C; IR (KBr): 1400 - 1600 (aromatic), 1645 (C=O),1659 (C=O), 2700 - 3200 (OH) cm-1; LCMS (ESI): m/z 306 [M-H]-; 1H-NMR (400 MHz, DMSO-d6): δ 7.32 (t, J = 8 Hz, 1H, quinoline H7), 7.39 (d, J = 8 Hz, 1H, quinoline H8), 7.47 - 7.49 ( m, 3H, benzylidene H3 & H4 & H5), 7.72 (t, J = 8 Hz, 1H, quinoline H6), 7.78 (m, 2H, benzylidene H2 & H6), 8.00 (d, J = 8 Hz, 1H, quinoline H5), 8.49 (s, 1H, = CH), 12.08 (s, 1H, NH), 13.33 (s, 1H, NH), 16.67 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 96.43, 114.57, 116.45, 123.12, 124.47, 128.03, 129.34, 131.14, 134.22, 137.73, 139.32, 151.33, 162.84, 167.95, 173.25; Anal. Calcd. for C17H13N3O3: C, 66.44; H, 4.26; N, 13.67; Found: C, 66.48; H, 4.31; N, 13.60.
2.6. 2-Chlorobenzylidene- 4-hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12b)
mp: 187°C; IR (KBr): 1400 - 1600 (aromatic), 1644 (C=O), 1669 (C=O), 2500 - 3300 (OH) cm-1; LCMS (ESI): m/z 340 [M-1]-; 1H-NMR (400 MHz, DMSO-d6): δ 7.33 (m, 1H, quinoline H7), 7.41 - 7.58 (m, 4H, quinoline H8 & 2-chlorobenzylidene H3 & H4 & H5), 7.74 (t, J = 8 Hz, 1H, quinoline H6), 8.04 (m, 2H, quinoline H5 & 2-chlorobenzylidene H6), 8.70 (brs, 1H, = CH), 12.10 (s, 1H, NH), 13.42 (s, 1H, NH), 16.52 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 94.60, 116.60, 118.09, 123.54, 124.65, 129.13, 129.94, 131.68, 134.50, 139.30, 141.09, 142.17, 142.38, 157.31, 163.22, 167.94, 172.83; Anal. Calcd. for C17H12ClN3O3: C, 59.75; H, 3.54; N, 12.30; Found: C, 59.71; H, 3.59; N, 12.33.
2.7. 3-Chlorobenzylidene- 4-hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12c)
mp: 198°C; IR (KBr): 1400 - 1600 (aromatic), 1658 (C=O),1683 (C=O), 2500 - 3300 (OH) cm-1; LCMS (ESI): m/z 340 [M-H]-; 1H-NMR (400 MHz, DMSO-d6): δ 7.33 (t, J = 8Hz, 1H, quinoline H7), 7.40 (d, J = 8Hz, 1H, quinoline H8), 7.49 - 7.53 (m, 2H, quinoline H6 & 3-clorobenzylidene H5), 7.71 - 7.76 (m, 2H, 3-chlorobenzylidene H4 & H6), 7.81 (s, 1H, 3-chlorobenzylidene H2), 8.01 (d, J = 8 Hz, 1H, quinoline H5), 8.49 (s, 1H, = CH), 12.11 (s, 1H, NH), 13.39 (s, 1H, NH), 16.54 (s, 1H, OH) ); 13C-NMR (100 MHz, DMSO-d6): δ 96.46, 114.52, 116.48, 123.19, 124.51, 126.58, 127.32, 130.76, 131.31, 134.10, 134.85, 136.44, 139.36, 149.78, 162.84, 168.12, 173.29; Anal. Calcd. for C17H12ClN3O3: C, 59.75; H, 3.54; N, 12.30; Found: C, 59.72; H, 3.57; N, 12.27.
2.8. 4-Chlorobenzylidene- 4-hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12d)
mp: 241°C; IR (KBr): 1400 - 1600 (aromatic), 1658 (C=O), 1683 (C=O), 2500 - 3300 (OH) cm-1; LCMS (ESI): m/z 340 [M-H]-; 1H-NMR (400 MHz, DMSO-d6): δ 7.33 (t, J = 8 Hz, 1H, quinoline H7), 7.39 (d, J = 8 Hz, 1H, quinoline H8), 7.54 (d, J = 8.4Hz, 2H, 4-chlorobenzylidene H3 & H5), 7.73 (t, J = 8 Hz, 1H, quinoline H6), 7.79 (d, J = 8.4Hz, 2H, 4-chlorobenzylidene H2 & H6), 8.00 (d, J = 8 Hz, 1H, quinoline H5), 8.49 (s, 1H, = CH), 12.10 (s, 1H, NH), 13.36 (s, 1H, NH), 16.60 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 96.44, 114.55, 116.47, 123.17, 124.49, 129.49, 129.63, 133.18, 134.82, 135.62, 139.35, 150.14, 162.84, 168.03, 173.27; Anal. Calcd. for C17H12ClN3O3: C, 59.75; H, 3.54; N, 12.30; Found: C, 59.79; H, 3.60; N, 12.26.
2.9. 2-Fluorobenzylidene- 4-hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12e)
mp: 195°C; IR (KBr): 1400 - 1600 (aromatic), 1647 (C=O), 1668 (C=O), 2660 - 3200 (OH) cm-1; LCMS (ESI): m/z 326 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 7.31 - 7.36 (m, 3H, quinoline H7 & 2-fluorobenzylidene H3 & H5), 7.40 (d, J = 8 Hz, 1H, quinoline H8), 7.51 - 7.57 (br s, 1H, 2-fluorobenzylidene H6), 7.73 (t, J = 8.4 Hz, 1H, 2-fluorobenzylidene H4), 7.95 (t, J = 8 Hz, 1H, quinoline H6), 8.01 (d, J = 8 Hz, 1H, quinoline H5), 8.60 (s, 1H, = CH), 12.09 (s, 1H, NH), 13.38 (s, 1H, NH), 16.56 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 94.58, 116.90, 118.13, 123.64, 124.55, 129.23, 130.04, 131.78, 134.60, 139.35, 141.04, 142.07, 142.37, 151.31, 158.09, 163.12, 167.84, 172.93; Anal. Calcd. for C17H12FN3O3: C, 62.77; H, 3.72; N, 12.92; Found: C, 62.72; H, 3.68; N, 12.99.
2.10. 3-Fluorobenzylidene- 4-hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12f)
mp: 237°C; IR (KBr): 1400 - 1600 (aromatic), 1648 (C=O), 1671 (C=O), 2800 - 3300 (OH) cm-1; LCMS (ESI): m/z 326 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 7.30 - 7.34 (m, 2H, quinoline H7 & 3-fluorobenzylidene H6), 7.39 (d, J = 8.4 Hz, 1H, quinoline H8), 7.51 - 7.57 (m, 2H, 3-fluorobenzylidene H2 & H5), 7.62 (d, J = 7.6 Hz, 1H, 3-fluorobenzylidene H4), 7.73 (t, J = 8.4 Hz, 1H, quinoline H6), 8.00 (d, J = 8.4 Hz, 1H, quinoline H5), 8.60 (s, 1H, = CH), 12.09 (s, 1H, NH), 13.38 (s, 1H, NH), 16.56 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 96.46, 113.88, 114.10, 114.52, 116.47, 117.79, 118.01, 123.18, 124.32, 124.50, 131.45, 131.53, 134.84, 136.70, 136.78, 139.35, 150.02, 161.57, 162.85, 163.99, 168.09, 173.28; Anal. Calcd. for C17H12FN3O3: C, 62.77; H, 3.72; N, 12.92; Found: C, 62.83; H, 3.64; N, 12.88.
2.11. 4-Fluorobenzylidene- 4-hydroxy- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12g)
mp: 212°C; IR (KBr): 1400 - 1600 (aromatic), 1649 (C=O),1667 (C=O), 2200 - 3400 (OH) cm-1; LCMS (ESI): m/z 326 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 7.30 - 7.35 (m, 3H, quinoline H7 & 4-fluorobenzylidene H2 & H6), 7.39 (d, J = 8.4 Hz, 1H, quinoline H8), 7.72 (t, J = 8.4 Hz, 1H, quinoline H6), 7.83 - 7.86 (m, 2H, 4-fluorobenzylidene H3 & H5), 8.00 (d, J = 8.4 Hz, 1H, quinoline H5), 8.50 (s, 1H, = CH), 12.09 (s, 1H, NH), 13.32 (s, 1H, NH), 16.65 (s, 1H, OH) ); 13C-NMR (100 MHz, DMSO-d6) : δ 96.41, 114.56, 116.37, 116.46, 116.59, 123.14, 124.47, 130.22, 130.31, 130.86, 134.75, 139.32, 150.25, 162.68, 162.84, 165.15, 167.94, 173.24; Anal. Calcd. for C17H12FN3O3: C, 62.77; H, 3.72; N, 12.92; Found: C, 62.71; H, 3.78; N, 12.96.
2.12. 4-Hydroxy-N'- (2-methylbenzylidene)- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12h)
mp: 203°C; IR (KBr): 1400 - 1600 (aromatic), 1632 (C=O),1658 (C=O), 2200 - 3200 (OH) cm-1; LCMS (ESI): m/z 322 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 2.50 (s, 3H, CH3), 7.27 - 7.37 (m, 4H, quinoline H7 & 2-methylbenzylidene H3 & H4 & H5), 7.40 (d, J = 8.4 Hz, 1H, 2- methylbenzylidene H6), 7.73 (d, J = 7.2 Hz, 1H, quinoline H8), 7.85 (t, J = 7.2 Hz, 1H, quinoline H6), 8.01 (d, J = 7.2 Hz, 1H, quinoline H5), 8.67 (s, 1H, = CH), 12.04 (s, 1H, NH), 13.26 (s, 1H, NH), 16.77 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 22.54, 94.65, 116.60, 118.09, 123.54, 124.60, 129.13, 129.83, 131.94, 134.38, 139.30, 141.09, 142.17, 142.50, 154.31, 163.12, 167.68, 172.94; Anal. Calcd. for C18H15N3O3: C, 67.28; H, 4.71; N, 13.08; Found: C, 67.22; H, 4.76; N, 13.01.
2.13. 4-Hydroxy-N'- (4-methylbenzylidene)- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12i)
mp: 181°C; IR (KBr): 1400 - 1600 (aromatic), 1662 (C=O), 2500 - 3300 (OH) cm-1; LCMS (ESI): m/z 322 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 2.35 (s, 3H, CH3),7.28 (d, J = 8.4 Hz, 2H, 4-methylbenzylidene H3 & H5), 7.32 (t, J = 7.6 Hz, 1H, quinoline H7), 7.39 (d, J = 7.6 Hz, 1H, quinoline H8), 7.67 (d, J = 8.4 Hz, 2H, 4-methylbenzylidene H2 & H6), 7.72 (t, J = 7.6 Hz, 1H, quinoline H6), 8.00 (d, J = 7.6 Hz, 1H, quinoline H5), 8.44 (s, 1H, = CH), 12.07 (s, 1H, NH), 13.27 (s, 1H, NH), 16.73 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 21.58, 96.41, 114.60, 116.45, 123.12, 124.47, 128.03, 129.95, 131.51, 134.72, 139.31, 141.11, 151.36, 162.84, 167.83, 173.23. Anal. Calcd. for C18H15N3O3: C, 67.28; H, 4.71; N, 13.08; Found: C, 67.33; H, 4.65; N, 13.15.
2.14. 4-Hydroxy-N'- (2-methoxybenzylidene)- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12j)
mp: 122°C; IR (KBr): 1400 - 1600 (aromatic), 1599 (C=O),1660 (C=O), 2700 - 3200 (OH) cm-1; LCMS (ESI): m/z 338 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 3.89 (s, 3H, OCH3), 7.05 (t, J = 7.6 Hz, 1H, 2-methoxybenzylidene H5), 7.13 (t, J = 8 Hz, 1H, quinoline H7), 7.32 (d, J = 8 Hz, 1H, quinoline H8), 7.40 (d, J = 7.6 Hz, 1H, 2-methoxybenzylidene H6), 7.47 (t, J = 8 Hz, 1H, quinoline H6), 7.73 (t, J = 7.6 Hz, 1H, 2-methoxybenzylidene H4), 7.87 (d, J = 7.6 Hz, 1H, 2-methoxybenzylidene H3), 8.00 (d, J = 8 Hz, 1H, quinoline H5), 8.58 (s, 1H, = CH), 12.08 (s, 1H, NH), 13.33 (s, 1H, NH), 16.67 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 56.24, 96.43, 111.44, 114.58, 116.45, 121.27, 121.94, 123.12, 124.46, 126.39, 132.84, 134.72, 139.30, 146.14, 158.57, 162.82, 167.79, 173.21; Anal. Calcd. for C18H15N3O4: C, 64.09; H, 4.48; N, 12.46; Found: C, 64.03; H, 4.44; N, 12.50.
2.15. 4-Hydroxy-N'- (3-methoxybenzylidene)- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12k)
mp: 190°C; IR (KBr): 1400 - 1600 (aromatic), 1641 (C=O), 1664 (C=O), 2600 - 3200 (OH) cm-1; LCMS (ESI): m/z 338 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 3.81 (s, 3H, OCH3), 7.02 (t, J = 8 Hz, 1H, quinoline H7), 7.29 - 7.41 (m, 5H, quinoline H8 & 3-methoxybenzylidene H2 & H4 & H5 & H6), 7.71 (t, J = 8 Hz, 1H, quinoline H6), 7.99 (d, J = 8 Hz, 1H, quinoline H5), 8.44 (s, 1H, = CH), 12.08 (s, 1H, NH), 13.33 (s, 1H, NH), 16.63 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 55.64, 96.44, 112.24, 114.56, 116.45, 117.27, 120.86, 123.14, 124.47, 130.46, 134.75, 135.59, 139.32, 151.22, 159.95, 162.85, 167.93, 173.23; Anal. Calcd. for C18H15N3O4: C, 64.09; H, 4.48; N, 12.46; Found: C, 64.04; H, 4.53; N, 12.51.
2.16. 4-Hydroxy-N'- (4-methoxybenzylidene)- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12l)
mp: 240°C; IR (KBr): 1400 - 1600 (aromatic), 1650 (C=O),1663 (C=O), 2500 - 3200 (OH) cm-1; LCMS (ESI): m/z 338 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 3.82 (s, 3H, OCH3), 7.07 (d, J = 8.4 Hz, 2H, 4-methoxybenzylidene H3 & H5), 7.32 (t, J = 8 Hz, 1H, quinoline H7), 7.39 (d, J = 8 Hz, 1H, quinoline H8), 7.70 - 7.75 (m, 3H, quinoline H6 & 4-methoxybenzylidene H2 & H6), 8.00 (d, J = 8Hz, 1H, quinoline H5), 8.41 (s, 1H, = CH), 12.06 (s, 1H, NH), 13.24 (s, 1H, NH), 16.80 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 55.82, 96.39, 114.63, 114.86, 116.44, 123.10, 124.45, 126.72, 129.75, 134.67, 139.27, 151.17, 161.78, 162.85, 167.64, 172.20; Anal. Calcd. for C18H15N3O4: C, 64.09; H, 4.48; N, 12.46; Found: C, 64.13; H, 4.44; N, 12.42.
2.17. 4-Hydroxy-N'- (2-hydroxybenzylidene)- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12m)
mp: 228°C; IR (KBr): 1400 - 1600 (aromatic), 1643 (C=O), 1663 (C=O), 2300 - 3300 (OH) cm-1; LCMS (ESI): m/z 322 [M-H]-; 1H-NMR (400 MHz, DMSO-d6): δ 6.92 (m, 2H, quinoline H7 & 2-hydroxybenzylidene H3), 7.29 - 7.36 (m, 2H, quinoline H8 & 2-hydroxybenzylidene H5), 7.39 (d, J = 8 Hz, 1H, quinoline H6), 7.58 (d, J = 7.6 Hz, 1H, 2-hydroxybenzylidene H6),7.72 (t, J = 7.6 Hz, 1H, 2-hydroxybenzylidene H4), 7.99 (d, J = 8Hz, 1H, quinoline H5), 8.68 (s, 1H, = CH), 11.00 (s, 1H, OH), 12.11 (s, 1H, NH), 13.36 (s, 1H, NH), 16.36 (s, 1H, OH). ); 13C-NMR (100 MHz, DMSO-d6): δ 96.39, 114.47, 116.47, 116.93, 118.86, 119.95, 123.13, 124.47, 130.10, 132.56, 134.78, 139.36, 151.25, 158.07, 162.77, 167.51, 173.04; Anal. Calcd. for C17H13N3O4: C, 63.16; H, 4.05; N, 13.00; Found: C, 63.12; H, 4.12; N, 12.89.
2.18. 4-Hydroxy-N'- (4-hydroxybenzylidene)- 2-oxo- 1,2-dihydroquinoline- 3-carbohydrazide (12n)
mp: 220°C; IR (KBr): 1400 - 1600 (aromatic), 1636 (C=O),1660 (C=O), 2600 - 3300 (OH) cm-1; LCMS (ESI): m/z 322 [M-H]-; 1H-NMR (400 MHz, DMSO-d6): δ 6.85 (d, J = 7.6 Hz, 2H, 4-hydroxybenzylidene H3 & H5), 7.31 (t, J = 8 Hz, 1H, quinolone H7), 7.39 (d, J = 8 Hz, 1H, quinolone H8), 7.63 (d, J = 7.6 Hz, 2H, 4-hydroxybenzylidene H2 & H6), 7.71 (t, J = 8 Hz, 1H, quinoline H6), 7.99 (d, J = 8 Hz, 1H, quinoline H5), 8.44 (s, 1H, = CH), 10.07 (s, 1H, OH), 12.04 (s, 1H, NH), 13.19 (s, 1H, NH), 16.86 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 96.35, 114.64, 116.20, 116.40, 122.99, 124.38, 125.14, 129.94, 134.52, 139.22, 151.39, 160.44, 162.82, 167.50, 173.14; Anal. Calcd. for C17H13N3O4: C, 63.16; H, 4.05; N, 13.00; Found: C, 63.10; H, 4.11; N, 13.04.
2.19. 4-Hydroxy-N'- (4-(methylthio) benzylidene)- 2-oxo- 1,2-dihydroquinoline- 3 -carbohydrazide (12o)
mp: 231°C; IR (KBr): 1400 - 1600 (aromatic), 1611 (C=O), 1671 (C=O), 2500 - 3200 (OH) cm-1; LCMS (ESI): m/z 354 [M+H]+; 1H-NMR (400 MHz, DMSO-d6): δ 2.52 (s, 3H, SCH3), 7.31 - 7.36 (m, 3H, quinolone H7 & 4-methylthiobenzylidene H3 & H5), 7.39 (d, J = 8Hz, 1H, quinolone H8), 7.71 (m, 3H, quinolone H6 & 4-methylthiobenzylidene H2 & H6), 8.00 (d, J = 8 Hz, 1H, quinolone H5), 8.44 (s, 1H, = CH), 12.08 (s, 1H, NH), 13.29 (s, 1H, NH), 16.72 (s, 1H, OH); 13C-NMR (100 MHz, DMSO-d6): δ 14.61, 96.43, 114.60, 116.46, 123.14, 124.48, 125.99, 128.42, 130.52, 134.74, 139.31, 142.38, 150.98, 162.84, 167.79, 173.23; Anal. Calcd. for C18H15N3O3S: C, 61.18; H, 4.28; N, 11.89; Found: C, 61.13; H, 4.31; N, 11.85.
2.20. Antibacterial Activity
The antibacterial activity of the compounds was evaluated by the broth microdilution method (21). The following strains were used in this study: Staphylococcus aureus PTCC 6538, Micrococcus luteus PTCC 9341 Bacillus cereus PTCC 6633, Escherichia coli PTCC 8739, Salmonella Typhi PTCC 14028, and Pseudomonas aeruginosa PTCC 9027. All strains were cultured in Soybean Casein Digest Agar (SCDA) and, after 24 hours of incubation, were diluted by 0.5 McFarland turbidity standards.
Different concentrations of the synthesized compounds (10 µL of each) were poured into the 96 well plates, to which80 µL of Muller Hinton Broth (MHB) medium and 10 µL of microbial suspensions were added. The final concentration of the microbial suspensions in each well was 1.5 × 107 cfu/mL. The plates were sealed to lower the solvent evaporation and then incubated at 35°C for 24 h. An ELISA reader spectrophotometer (TECAN-SP) was used to read the optical density of the wells at 580 nm. The inhibitory concentration (IC) in each well was measured by the following equation:
In this equation, ODa, ODb, and ODc determine the optical density of the solutions containing microorganisms and test compounds, only test compounds, and only microorganisms, respectively. Moreover, IC50 is defined as the lowest concentration of the test compound, at which the bacterial growth was disrupted. The standard antibiotics were ciprofloxacin and nalidixic acid. Each assay was performed as duplicates.
2.21. Molecular Docking Study
Autodock Vina software was used to perform a molecular modeling study (22). In this study, 3OYA was used to analyze the binding mode of the compounds in the IN active site. Autodock tools 1.5.6 from the MGL Tools package were utilized to prepare the protein and ligands’ structures (23). First, the co-crystallized raltegravir and water molecules were removed from the protein structure. Then Kollman charges were calculated, nonpolar hydrogens were removed, and AutoDock4 atom type was assigned to the protein structure. HyperChem 8.0 was used to create and optimize the ligand molecule (24). The Grid box with 20 × 20 × 20 dimensions was defined around the crystallographic ligand, raltegravir, and regarded as the active site. Autodock Vina was used to dock the molecule in the active site and produce the bioactive conformations.
3. Results and Discussion
3.1. Chemistry
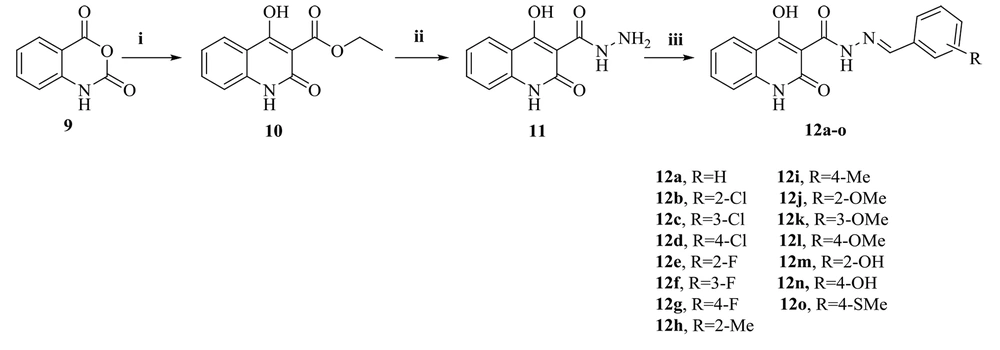
Figure 3 shows the synthesis path of the target compounds (12a-o). Isatoic anhydride (9) was the starting material for the reaction with diethyl malonate in dimethylformamide (DMF) as a solvent. This process afforded the expected ethyl 4-hydroxy-2-oxo-1,2-dihydroquinoline-3-carboxylate (10), whose reaction with hydrazine hydrate provided the corresponding carbohydrazide intermediate (11). Finally, the target compounds (12a-o) were obtained during the reaction of the compound (11) with benzaldehyde derivatives in acceptable yields. IR, 1H-NMR, 13C-NMR spectroscopy, and LC-MS were used to confirm the structure of all synthesized derivatives.
3.2. Anti-HIV-1 Activities
A series of N'-arylidene-4-hydroxy-2-oxo-1,2-dihydroquinoline-3-carbohydrazide derivatives (12a-o) were synthesized and evaluated in vitro regarding the disruption of both IN ST process and the cell-based HIV-1 replication, according to the previously reported procedures (25-28). The positive control was raltegravir. The cytotoxicity of the synthesized compounds was also assayed by a cell-based MTT method. The biological activity of the compounds is represented as IN IC50, anti-HIV-1 EC50, and CC50 in Figure 4.
The results revealed that all tested compounds exhibited no cytotoxicity at concentrations < 250 µM. Accordingly, this scaffold would provide a safe template for the anti-HIV-1 drug design. In integrase enzymatic assay, the compounds displayed no significant inhibitory activity at concentrations < 100 µM. Moreover, low antiviral activity was observed in the cell-based anti-HIV-1 assay. This might be due to poor permeability or physicochemical properties.
3.3. Antibacterial Activities
The final compounds (12a-o) were also tested for their in vitro antibacterial activities toward three Gram-positive bacterial species (namely S. aureus PTCC 6538, M. luteus PTCC 9341, and B. cereus PTCC 6633) and three Gram-negative bacterial species (namely E. coli PTCC 8739, S. Typhi PTCC 14028, and P. aeruginosa PTCC 9027) using the minimum inhibitory concentration (MIC) assay. The positive controls were ciprofloxacin and nalidixic acid; however, 10% dimethyl sulfoxide (DMSO) in water was used as the negative controls. The negative control had no impact on the antibacterial activity. The selected antibacterial efficacy results of the tested compounds are presented in Table 1.
The analysis of the antibacterial results revealed that only compounds 12a, 12b, 12j, and 12n exhibited MIC < 100 µg/mL. Compound 12a with a phenyl group was potent against S. aureus and E. coli with MIC = 78 µg/mL. Compound 12n with a 4-hydroxyphenyl group had the same MIC values (= 78 µg/mL) against S. aureus, B. cereus, E. coli, and P. aeruginosa. Compound 12j with a 2-methoxyphenyl group showed an acceptable activity (MIC = 78 µg/mL) only against M. luteus. The best antibacterial activity was demonstrated by the compound possessing a 2-chlorophenyl group, 12b with MIC = 39 µg/mL against S. aureus, E. coli, and P. aeruginosa. The estimated MIC values suggest that the designed compounds can be used for further development by structure modification to discover more active molecules.
| Compounds | Gram-Positive Bacteria | Gram-Negative Bacteria | ||||
|---|---|---|---|---|---|---|
| Staphylococcus aureus | Bacillus cereus | Micrococcus luteus | Escherichia coli | Pseudomonas aeruginosa | Salmonella Typhi | |
| 12a | 78 | > 1000 | > 1000 | 78 | 156 | 312 |
| 12b | 39 | 156 | 156 | 39 | 39 | 156 |
| 12c | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 |
| 12d | 625 | > 1000 | > 1000 | > 1000 | - | > 1000 |
| 12e | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 |
| 12f | - | 625 | - | - | - | - |
| 12g | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 |
| 12h | 312 | > 1000 | > 1000 | 625 | - | - |
| 12i | > 1000 | > 1000 | > 1000 | > 1000 | - | - |
| 12j | > 1000 | 156 | 39 | > 1000 | > 1000 | - |
| 12k | 312 | - | - | > 1000 | - | - |
| 12l | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 |
| 12m | - | - | - | > 1000 | - | - |
| 12n | 78 | 78 | - | 78 | 78 | - |
| 12o | > 1000 | > 1000 | 625 | > 1000 | 625 | > 1000 |
| Ciprofloxacin | 1.92 | 3.92 | 1.95 | 62.5 | ||
| Nalidixic acid | 3.92 | 1.95 | 3.92 | 15.62 | ||
| DMSO | - | - | - | - | - | - |
Selected MIC (µg/mL) Results for Synthesized Compounds
3.4. Molecular Modeling
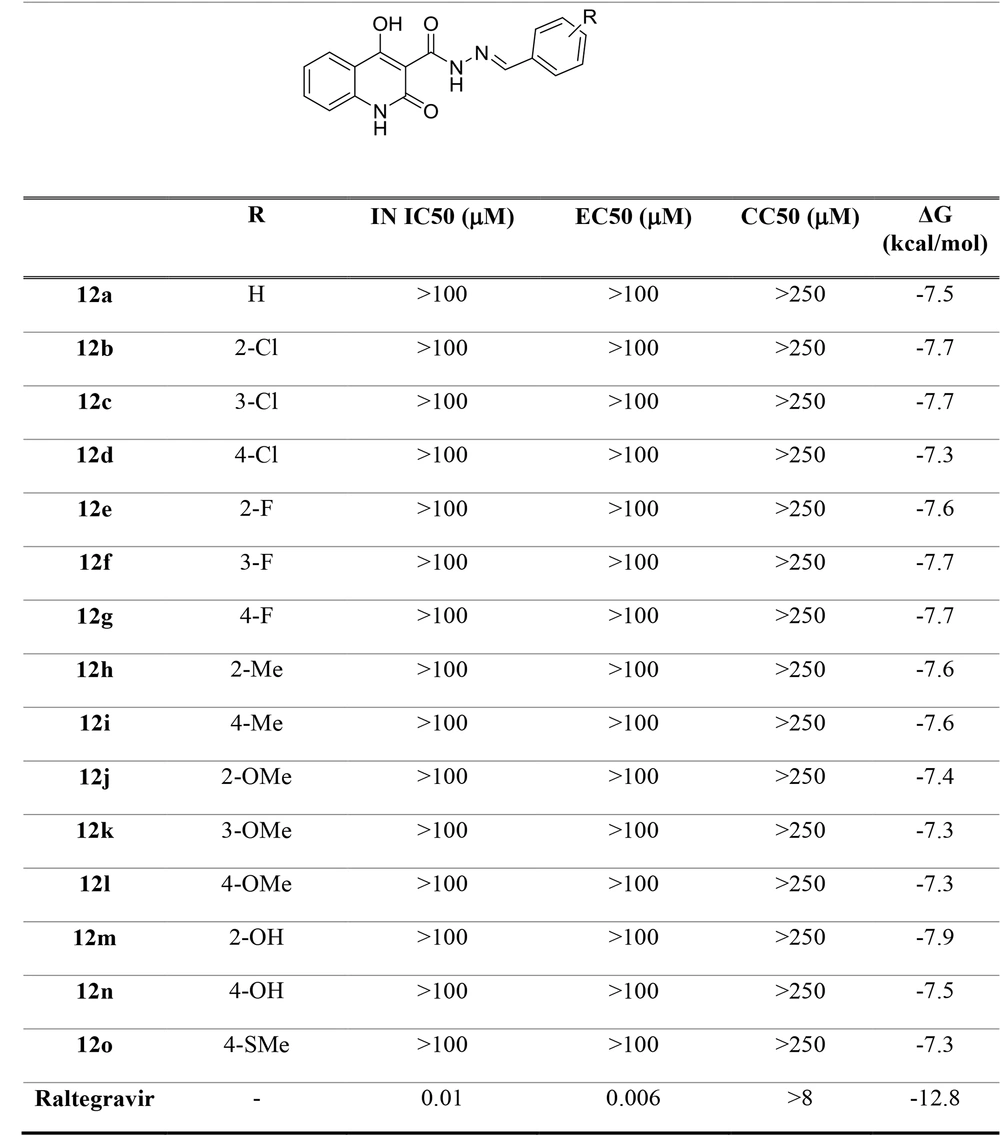
In this study, N'-arylidene-4-hydroxy-2-oxo-1,2-dihydroquinoline-3-carbohydrazide derivatives were assumed as potential new IN inhibitors. The hypothesis was then examined using a computational docking study. In the docking study, the receptor was the prototype foamy virus integrase (PFV-IN) structure encompassing two Mg2+ ions and a double chain DNA at 2.65 Å resolution (PDB:3OYA) (29, 30). The docking study was performed by a flexible-ligand and rigid target docking experiment using Autodock Vina software. It was then validated by redocking the co-crystalized ligand, raltegravir, under the same condition and superimposition on the co-crystallized ligand pose (RMSD = 0.001). Raltegravir revealed high-affinity binding (-12.8 kcal/mol) to the IN active site.
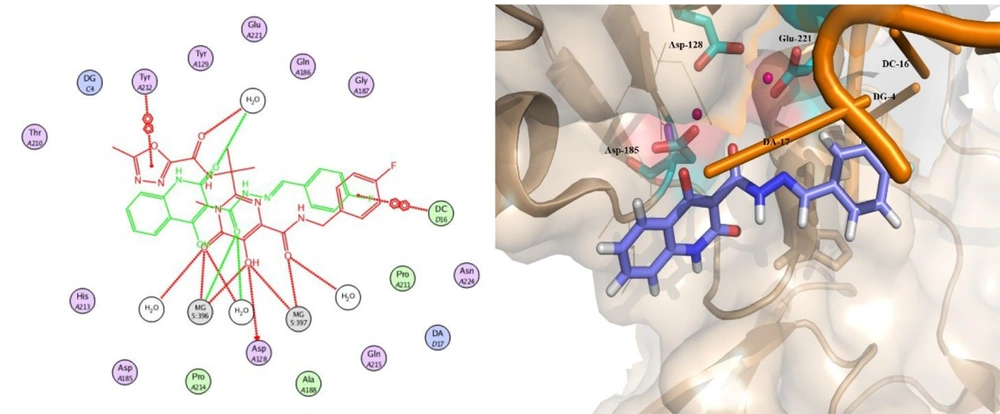
Docking studies demonstrated that all docked compounds occupied a nearly identical location in the integrase active site. The affinity binding energy of compounds ranged from -7.3 to -7.9 kcal/mol (Figure 4). Figure 5 shows the 2D and 3D alignment of one of the designed compounds, i.e., 12g, in the active site. As presented in Figure 5, the hydroxy and carboxamide groups of Compound 12g interacted with the Mg2+ ion, as expected. Moreover, the p-fluorophenyl group fit into a tight pocket created by guanine 4 (DG4), cytosine 16 (DC16), and adenine 17 (DA17).
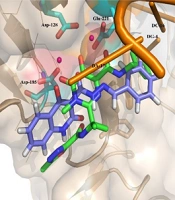
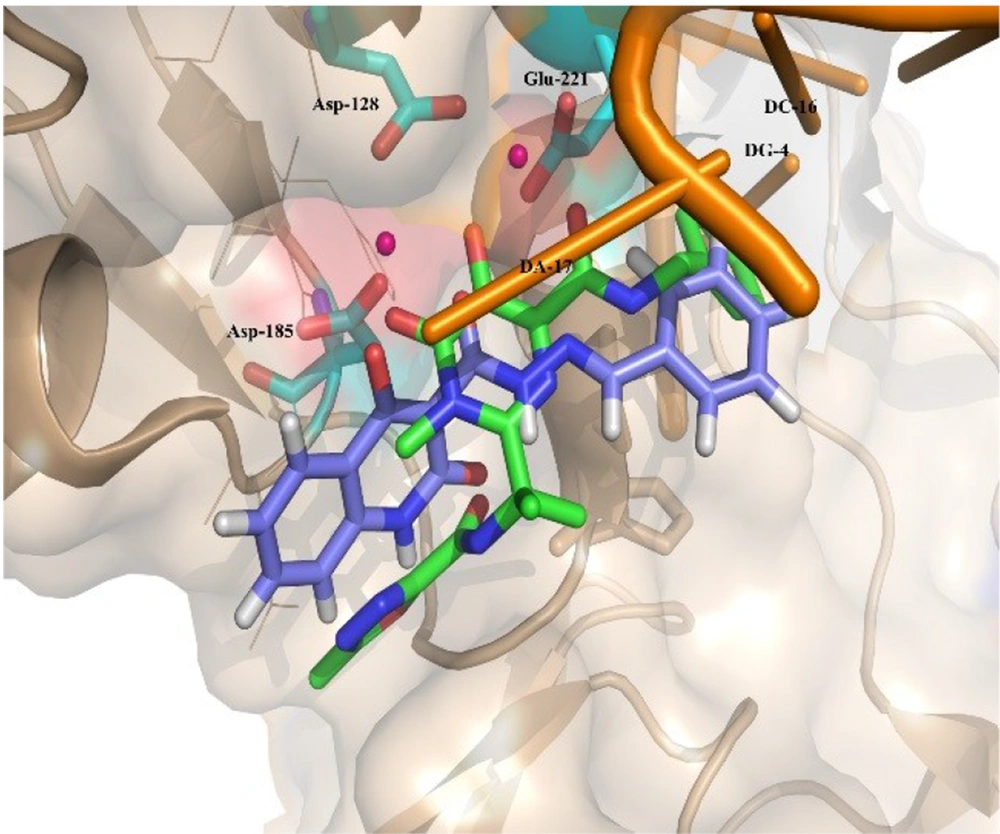
Moreover, Figure 6 shows that the binding pose of Compound 12g is similar to that of Raltegravir. In general, the binding mode of the docked compounds was similar to raltegravir. However, the designed compounds showed no significant anti-HIV activity, which may be due to unfavorable physicochemical properties.
3.5. Conclusions
A series of novel N'-arylidene-4-hydroxy-2-oxo-1,2-dihydroquinoline-3-carbohydrazide derivatives were designed and synthesized and were then evaluated in terms of anti-HIV-1 and antibacterial activities. The designed compounds revealed no significant anti-HIV-1 activity at concentrations < 100 µM. In an in vitro antibacterial assay using the MIC method, the best activity was observed by Compound 12b, which exhibited the MIC value of 39 µg/mL against S. aureus, E. coli, and P. aeruginosa. These findings indicated that the 4-hydroxy-2-oxo-1,2-dihydroquinoline-3-carbohydrazide scaffold provides an acceptable chemical template for synthetic modification, probably resulting in compounds with further promising anti-HIV-1 and antibacterial potencies.