1. Background
Infant formula is food, and breast milk is given as slurry to babies from four to six months (1, 2). Most infants use cereal-based complementary foods prepared at the household level (3, 4). Genetically modified crops (GMCs) cultivated in 28 countries had reached 2.001 million km2, among which maize and soybean accounted for 30.73% and 50% of these crops (5). North America, India, and some countries in Continental America are the top GMCs growing countries, producing 91.3% of global GMCs in 2020 (6).
One of the most used staple food and the second most cultivated GMC with the highest number of approvals is maize (ISAAA, 2020). The most common GM plants are maize, soybean, rapeseed (canola), and cotton (7, 8). There are concerns about the risks to biodiversity and the environment, providing information to the consumers. Food labels are regarded as mandatory by the European Union for products above 0.9% GM (9). DNA-based methods, such as polymerase chain reaction (PCR), have been the technique of choice for analyzing and detecting GM organisms (GMOs) in foods. Real-time PCR is used for identifying and quantifying transgenic products by the European Union. Today, PCR has been proven sensitive, precise, and accessible for detecting transgenic organisms (10).
Resistance to herbicides is the main characteristic of GMCs, and 47% of the world has planting areas of herbicide-resistant corn, soy, cotton, and canola. The usage of these products can significantly save on pesticides and herbicides, making the environment less contaminated (11). There are many reports from Iran (12), Serbia (13), and Italy (14) that GMOs are present in various foods. The endogenous gene for soybean is lectin and for maize is zein (15). The pat gene expression causes tolerance to the herbicide phosphinothricin in transgenic plants (16). Often NOS terminator is used as an insert for GMOs and a biomarker of transgenic plants (17). As mentioned above, NOS terminator and pat are the most found elements in approved transgenic products worldwide (18). The NOS terminator DNA commonly used in GMCs does not exist in the genomes of ordinary plants (19). One of the most widely used elements that confer gluten tolerance to the recipient plant for generating GM is the pat gene (20). Except for regulating the labeling, there are no specific rules to control the presence or use of GMOs in products (21). In some countries, consumers' freedom of choice is ensured by labels on all GMO-derived foods (14).
2. Objectives
GMOs are prohibited from being used in baby food and infant formula. Therefore, this study aimed to detect the NOS terminator and pat genes in GM soybean and maize as common genes in GMO products. The real-time PCR method using specific primers was applied to commercially available infant formula and baby food in Iran.
3. Methods
3.1. Samples
A total of 30 baby food (30 samples of 15 various brands) and 30 infant formula (30 cases of 10 different brands) suitable for children under 12 months were taken. Samples were randomly collected from a local drugstore in Iran in 2019. The samples comprised 30 different brands produced by 12 manufacturers from six countries. We chose these samples due to containing soybean and maize, which are possible to have GMO.
3.2. Reference Materials
Certified reference materials (CRMs) for positive and negative controls provide permission to validate analytical procedures (10). Maize powder containing 0.1%, 0.5%, 1%, and 5% of MON810 maize and soybean (A2704-12) was used. In addition, we utilized GM-free soy and maize powders as the negative control. All CRMs were prepared from the Joint Centre, Institute for Reference Materials and Measurements (JRC-IRMM), Geel, Belgium.
3.3. DNA Extraction
Samples and certified reference material genomic DNA were extracted by PureLink Genomic Plant DNA Purification Kit (Thermo Fisher, USA). Afterwards, the DNA specimens were kept at -20°C until analysis. After extraction, DNA was quantified by spectrophotometry using a Thermo Scientific NanoDrop.
3.4. Oligonucleotide Primers
Primer pairs (LEC-F/R and ZE-F/R) were used in this study to measure and assess the quality of extracted DNA from baby food, infant formula, soy CRM, and maize CRM. We considered lectin and zein genes for primer designing based on the published papers (22, 23). According to the international standard management (ISO 21569: 2005), the primer pair pat and NOS terminator were prepared (24). In order to detect NOS terminator and pat, gene-specific primers were used for the event-specific detection (A2704-12, MON810) events using real-time PCR. The detection of MON810 and A2704-12 events was accomplished using the primer sets PA-R/PA-FA and NOS-F/NOS-R, respectively. The primers were diluted to a final concentration of 10 µM with sterile double distilled water and were stored at -20°C until use. The maize and soybean primers targeting transgenics are presented in Table 1.
| Target | Primer Sequence (5′ - 3′) | Amplicon (bp) | Reference |
|---|---|---|---|
| Maize zein | 139 | (22) | |
| ZE-F | CGCCAGAAATCGTTTTTCAT | ||
| ZE-R | GGTGGTGTCCTTGCTTCCTA | ||
| NOS terminator | 84 | (25) | |
| NOS-F | CGATGACGTTATTTATGAGATGGG | ||
| NOS-R | GACACCGCGCGCGATAATTTATCC | ||
| Pat | 108 | (26) | |
| PA-F | GCA AAA AAG CGG TTA GCT CCT | ||
| PA-R | ATT CAG GCT GCG CAA CTG TT | ||
| Soybean lectin | 110 | (23) | |
| LEC-F | AGTCTCATTCTTCTTTTTCCTGCTA | ||
| LEC-R | ATGGTGAATAATAAGCAGTGTAGATTG |
3.5. Real-time PCR Conditions
In order to verify and estimate the contents of maize and soybean detected by real-time PCR, the double calibration curve was plotted for the two identified events (MON810, A2704-12). The quantitative PCR assays were carried out in 20 μL of total reaction volume containing 0.5 μL of DNA extract (40 - 50 ng/μL), 0.5 μL of each forward and reverse primer (10 pmol), 10 μL of ready to use cyber green master mix (Amplicon, Denmark) (including Tris-HCl pH 8.5, 3 mM MgCl2, 0.4 mM dNTPs, (NH4) 2SO4, 0.2% Tween 20, 0.2 units/μL Amplicon Taq DNA polymerase, and 9 μL of ultrapure water. The real-time PCR program for pat and NOS was denaturation for 10 min at 95°C, amplification for 15 sec at 95°C, 60 sec at 59°C, and 30 sec at 72°C, 40 cycles, and final extension for 5 min at 72°C. For zein and lectin, the program was denaturation for 10 min at 95°C, amplification for the 30 sec at 94°C, 30 sec at 60°C, 25 sec at 72°C, 35 cycles, and final extension for 7 min at 72°C.
4. Results and Discussion
We collected and analyzed 60 infant formula and baby food containing soy and maize. All samples were assessed for internal detection control (zein and lectin) and transgenic sequences (pat, NOS terminator) by real-time PCR. The absorbance of DNA obtained from samples in this study had a range of 1.7 - 2 at 260 nm, and the concentration of DNA was 80 - 100 ng/μL. Similarly, other studies showed the possibility of extracting high-quality DNA from raw and middle processed food products by the exact extraction method (27).
4.1. Screening and Event-Specific Detection
To detect GM sequences, real-time PCR was applied with soy and maize CRMs containing various GM levels (0.1%, 0.5%, 1%, and 5%). In the first step, the positive result for endogenous genes, lectin, and zein, in all samples showed the validity of the DNA extraction method and the presence of high-quality DNA to be used in real-time PCR for the baby food and infant formula. Based on Results of screening for the target genes (pat and NOS terminator), amplification was only observed in CRMs for NOS and pat gene in three baby food samples and three infant formula. Samples numbers 20 and 25 out of Iranian and sample 25 among the imported baby food were positive. In addition, samples 52 and 57 among the imported and 40 in Iranian infant formulas were positive (Table 2). The results of baby food samples 4, 10, and 25 and infant formulas 35, 48, and 54 are listed in Table 2. Results with Ct > 40 were considered negative and are not included in Table 2.
| Sample | DNA Concentration (ng/μL) | Pat Gene CT Values | CRM Mon810 CT | NOS Terminator CT | CRM A2704-12 CT |
|---|---|---|---|---|---|
| 4 | 180.5 | 29.48 | 22.98 | Non detected | 18.76 |
| 20 | 176.8 | 30.70 | 23.65 | Non detected | 19.63 |
| 25 | 165.8 | 31.92 | 23.89 | Non detected | 20.87 |
| 48 | 134.9 | 29.95 | 24.32 | Non detected | 20.95 |
| 52 | 190.7 | 30.36 | 25.34 | Non detected | 19.43 |
| 57 | 116.1 | 29.63 | 23.63 | Non detected | 20.86 |
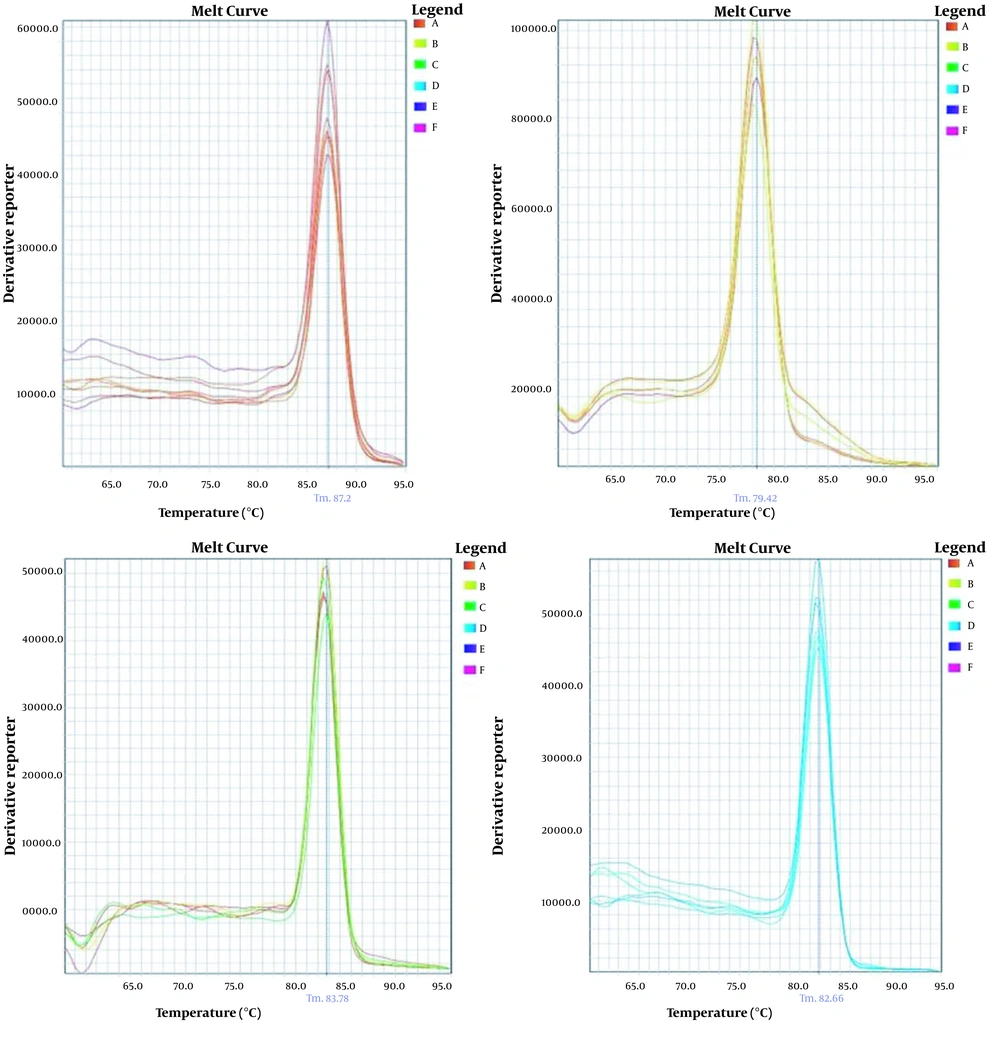
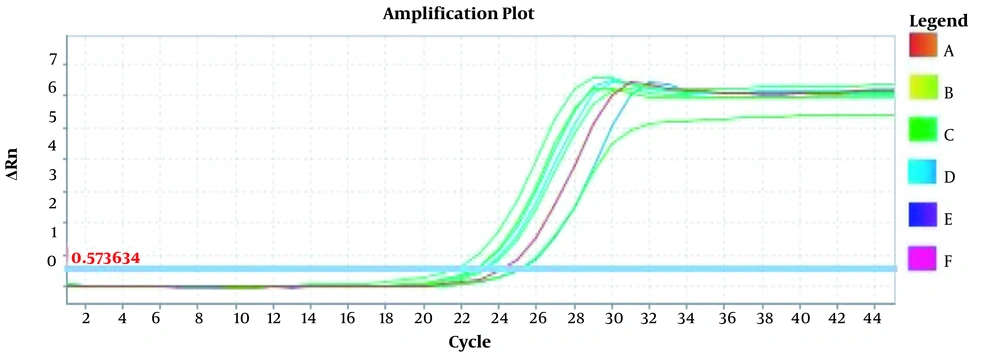
In order to analyze the specificity of real-time PCR for the products and the absence of unspecific PCR products and dimer primers, the melt curves of each gene were drawn separately (Figure 1).
The second step was the detection of pat and NOS terminator genes as GMO targets. Although the present study indicated that 5% of baby food and 5% of infant formulas were positive for target pat, none samples were positive for NOS terminator (Table 3).
| Sample Type | Number of Samples | Pat Positive | NOS Positive | Endogenous Gene: Lectin | Endogenous Gene: Zein | Negative |
|---|---|---|---|---|---|---|
| Baby food (Iranian) | 15 | 2 | Non detected | 15 | 15 | 13 |
| Baby food (imported) | 15 | 1 | Non detected | 15 | 15 | 14 |
| Infant formula (Iranian) | 15 | 1 | Non detected | 15 | 15 | 14 |
| Infant formula (imported) | 15 | 2 | Non detected | 15 | 15 | 13 |
| Total | 60 | 10% | 0% | 100% | 100% | 90% |
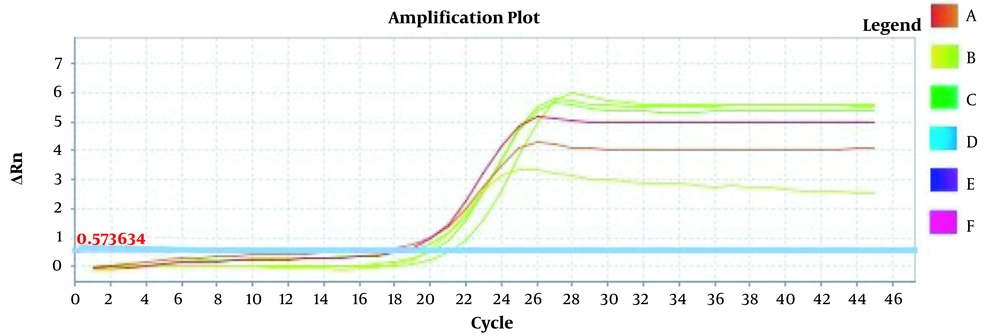
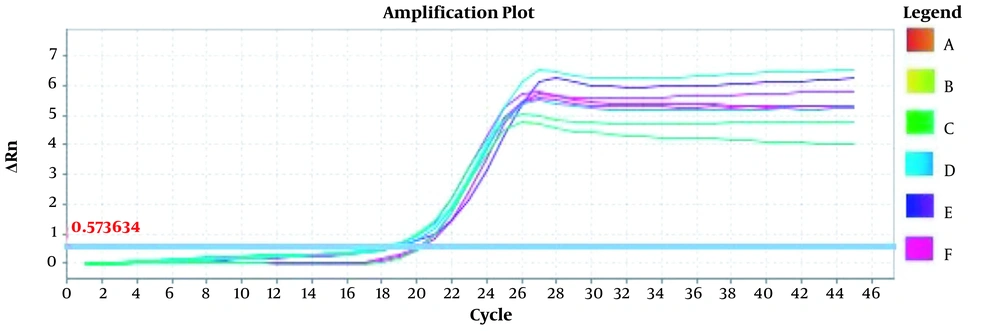
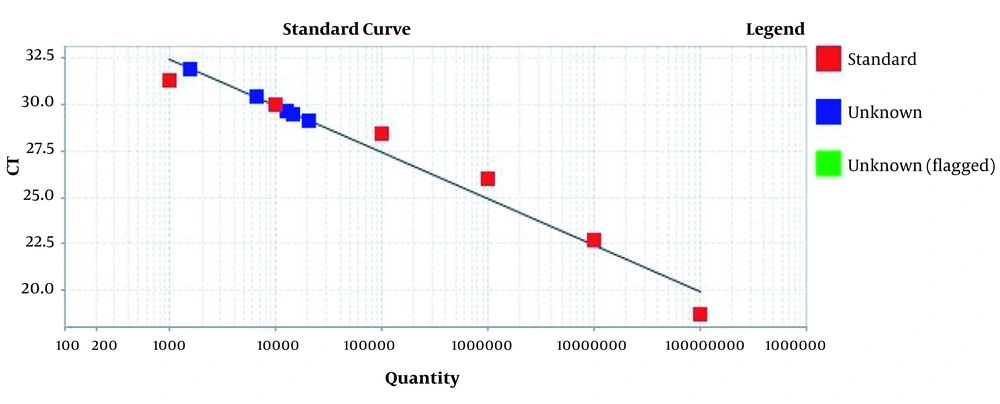
Amplification plots for CRM Mon810, CRM A2704-12, and pat gene detection in real-time using the ABI and standard curve for the pat gene (Figures 2 - 5). The standard curve was obtained from the data given in the equation and the fit of the lines.
Previous studies (9, 18) showed that DNA is seriously damaged during processing, making it difficult to amplify DNA fragments from processed samples. The research demonstrated a highly efficient DNA extraction method for processed foods, including maize-derived products, for GMO analysis (22). We collected samples like the results of Safaei et al. Investigations revealed that 91.2% of unlabeled soy products from the Hungarian market contained 35 S-promoter or NOS-terminator factors in their genome (28). Pat and NOS terminator genes are found in commercially transgenic plants and are detected in maize and soybean by real-time PCR (29). Two hundred cases, including maize, soy, and rice, were evaluated in another study to find GM (30).
Furthermore, a study in Turkey showed that baby food was not transgenic (31), while in our study, six out of 60 samples were positive for the pat gene. In another study in Turkey, 57 unlabeled products were evaluated for GM targets, and 11 of them tested positive for unlabeled GM products (32). Previous research in Iran showed that more than half of soybeans imported from other countries are transgenic. In this study, the sensitivity of the method was determined using acceptable CRMs (21). Elsanhoty et al. in Saudi Arabia found that 44% of randomly collected corn samples contained NOS and 35S (30).
A study conducted in Iran in 2020 on the middle-processed soy and maize samples indicated that 95% of soybeans and 60% of maize samples had a 35S promoter and NOS terminals (28). These factors also occur naturally in various plants and soil microorganisms. Therefore, GM sequences are probable in food products (29). In the research by Özgen Arun et al., screening of processed foods by detecting NOS terminator showed that 25% of the samples were GM-positive (32). Oraby et al. reported that 12.5% of CaMV 35S foods were positive, while the NOS terminator was negative in the same samples (33).
A study in Portugal by Fernandes et al. reported that the results of GMO screening on infant formula were 0% for 35S promoter and NOS terminator, while results vary with different samples (22). In our study, 10% of baby food and infant formula samples were pat. As shown in Table 3, further evaluation revealed that all six GM-positive samples contained the pat gene, proving GM sequences in their genome. Most of the positive GM samples came into Iran from other countries. This article also provides specific genes to detect the endogenous targets (i.e., lectin and zein for soybean and maize, respectively). The study results demonstrated that 6 out of 60 samples were positive for pat. Özgen Arun et al. showed that 8 of 14 tested samples contained Bt11 maize transgenic (32). Moreover, another investigation in Iran revealed that CaMV35 S, Bt11, and MON810 promoters were present in maize (34).
4.2. Conclusions
According to the results of the present study, we concluded that real-time PCR could be a suitable method for detecting GMOs even in highly prepared food products, such as baby food and infant formula. However, despite the current rules, consumers do not include the GMO type on the labels of products. Therefore, tracing the GM materials within baby food and infant formulas is vital for controlling inclusion and ensuring correct labeling.