1. Background
Although conventional cancer treatment strategies, such as surgery, chemotherapy, and radiotherapy, have demonstrated promising efficacy, their nonspecific activities and adverse side effects on both normal and tumor cells remain controversial (1). The blockade of signaling pathways with small-molecule inhibitors (SMIs) has represented a novel promising strategy for cancer treatment. Small molecule drugs are much smaller than monoclonal antibodies (mAbs) and can easily target extracellular molecules, cell surface ligand-binding receptors, and intracellular proteins (2). The SMIs are simple to use and can be utilized in conjunction with other therapies, such as chemotherapy, radiotherapy, immunotherapy, ribonucleic acid (RNA) interference molecules, and chemical-biology probes (2). Various SMIs have been currently investigated in clinical trials for hematopoietic and solid malignancies (3, 4).
In breast cancer, increasing evidence suggests that tumor-intrinsic signaling pathways regulate the immunosuppressive microenvironment and tumor immune escape (5). The phosphoinositide 3-kinase (PI3K)/protein kinase B (also known as AKT)/mammalian target of rapamycin (mTOR) signaling cascade is one of the most well-characterized signaling networks in normal and tumor cells (1). This network regulates cell fate by controlling the cell cycle, proliferation, glucose metabolism, and protein synthesis (3). Additionally, this pathway serves as a focal point of convergence for various other oncogenic activating signals. The constitutive activation of the PI3K/AKT/mTOR pathway is necessary for breast tumorigenesis (6, 7). Previous research has indicated that the PI3K/AKT/mTOR pathway is hyperactive in hormone receptor-positive breast cancers (8-11). Furthermore, Bruton’s tyrosine kinase (BTK) is a critical mediator associated with the PI3K/AKT/mTOR pathway that inhibits cancer cells from apoptosis in patients with breast cancer (12, 13). Moreover, it has been reported that there is a correlation between the immune evasion of cancer cells and the overactivation of this pathway (14).
Among the numerous mechanisms involved in the suppression of immune responses and surveillance of cancer cells, the expression of programmed death-ligand 1 (PD-L1) is modulated via the PI3K/AKT/mTOR and BTK signaling pathways. The PD-L1 plays a critical role in various cancer types, including chronic lymphocytic leukemia and lung carcinoma (15, 16). This immune checkpoint ligand is expressed on immune cells and different cancerous cells, including those detected in estrogen-positive breast cancers (17). The expression of PD-L1 as an immune evasion marker is regulated by PI3K/AKT/mTOR signaling elements or BTK-dependent signals. It is well established that the interaction between PD-L1 and its receptor PD-1 regulates the exhaustion of chronically stimulated T cells (18). Along with PD-L1, cluster of differentiation 155 (CD155) is another immune checkpoint ligand that is used as an immunomodulatory marker in hormone receptor-positive breast cancers (19). As with PD-L1, CD155 is highly expressed in cancer cells in the tumor microenvironment (20), and it has been found that its expression is induced through the signals sent by the AKT/mTOR cascade.
Furthermore, galectin-9 (Gal-9) is an immunoregulatory ligand that interacts with T-cell immunoglobulin mucin-3 (TIM-3) (5). The TIM-3 is a surface immune checkpoint protein that is considered a key molecule attributed to the dysfunctional state of T cells and the induction of immunological tolerance (21-23). It is well established that impaired natural killer (NK) and T-cell responses and tumor progression occur due to the engagement of immune checkpoint molecules to their associated receptors on T cells (24, 25).
Dysregulated signaling pathways, such as AKT-mTOR and BTK, are activated in breast cancer. Additionally, it is critical to recognize that immune checkpoint ligand expression regulation is highly complicated and highly dependent on the state of underlying signaling pathways. Intriguingly, most studies in this field focus on PD-L1 expression, with relatively limited attention paid to other checkpoint ligands, such as Gal-9 and CD155.
2. Objectives
Given the success of SMI-targeted therapy and immune checkpoint blocking strategy in breast cancers, this study investigated the associations between three SMIs and the immune evasion mechanisms of breast cancer cells.
3. Methods
3.1. Cell Culture
The human breast cancer cell line MCF-7 (NCBI code: C135) was obtained from the Iranian National Cell Bank and used in this study as a model of breast cancer. The cells were cultured in RPMI 1640 medium (Biowest, USA) supplemented with 10% heat-inactivated fetal bovine serum (Biowest, USA), 100 IU/mL penicillin (Biowest, USA), and 100 μg/mL streptomycin (Biowest, USA). The cells were incubated at 37°C in a humidified incubator containing 5% CO2 (Binder, Germany).
3.2. Half Maximal Inhibitory Concentration Determination of Small-Molecule Inhibitors
Three SMIs, including everolimus (mTOR inhibitor), MK-2206 (AKT inhibitor), and ibrutinib (BTK inhibitor), were applied in this study (Cayman, USA). These chemical reagents were dissolved in dimethyl sulfoxide (DMSO) (Merck, Germany), and aliquots were frozen. The MCF-7 cells were seeded in 96-well culture plates (SPL, South Korea) in a 200 μL fully supplemented medium in the presence of various drugs at concentrations ranging from 16 to 8192 nM and incubated for 48 hours to determine the half-maximal inhibitory concentration (IC50) values for all SMIs. Subsequently, each well was filled with 20 μL of freshly prepared 5 mg/mL of MTT solution (Sigma-Aldrich, USA) and incubated at 37 °C for 4 hours. Following incubation, the medium was carefully removed, and the reduced formazan crystals were dissolved in 150 μL DMSO by incubating at 37°C for 30 minutes in the dark. Optical density (OD) values were measured at 570 and 630 nm using an enzyme-linked immunosorbent assay plate reader (Synergy H1 BioTek, USA). All the experiments were performed in triplicates. The relative cell proliferation was calculated by dividing the mean OD values of each group by the OD values obtained from the control group.
3.3. Western Blot Analysis
For the determination of whether the selected SMIs affect signal transducer and activator of transcription 3 (STAT3)-Ser727 phosphorylation, exponentially growing cells were treated for 48 hours with an optimized concentration of SMIs and lysed to prepare protein extracts. The cells were homogenized for 30 minutes in ice in a RIPA buffer (Santa Cruz Biotechnology, USA) containing 150 mM NaCl, 1% NP-40, 50 mM Tris-HCl (pH 7.4), 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), protease inhibitor 1:50, and phenylmethylsulfonyl fluoride 1: 100, sodium orthovanadate 1: 100, and phosphatase inhibitor cocktail (Sigma-Aldrich, USA). After centrifugation at 10000 g for 10 minutes at 4°C, the protein samples were collected, and supernatants were stored at -80°C. The lysate was electrophoresed in a 10% SDS-polyacrylamide gel to separate the protein mixtures (Bio-Rad, UK) and then blotted onto a polyvinylidene difluoride membrane (Merck Millipore, Germany). For blocking, the membrane was dissolved in 4% bovine serum albumin (Sigma-Aldrich, USA) in Tris-buffered saline containing 0.05% Tween-20 (TBST) at 4°C for 45 minutes. After three washes with TBST washing buffer, the membranes were incubated overnight at 4°C with a specific rabbit anti-human phosphorylated STAT3 (p-STAT3) antibody (Cell Signaling, USA). Then, incubation was performed with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Sigma-Aldrich, USA) at room temperature for 1 hour. For semiquantitative analysis, a mouse antibody to β-actin (Santa-Cruz, USA) was used as internal control with an appropriate secondary HRP-conjugated goat anti-mouse antibody (Sigma-Aldrich, USA). Finally, antigen-antibody interactions were detected with enhanced chemiluminescence reagent according to the manufacturer’s specifications (ELC kit: CytoMatin Gene, Iran) and analyzed using the G: BOX Chemi XRQ system (Syngene, USA).
3.4. RNA Isolation and cDNA Synthesis
Following the MCF-7 cell line culture with various drugs, total RNA was isolated using the RNeasy kit (CinnaGen, Iran) according to the manufacturer’s instructions. The quality of the extracted RNA was confirmed by a nanospectrophotometer (WPA, England) and electrophoresis (Bio-Rad, UK). Complementary deoxyribonucleic acid (cDNA) was reverse-transcribed from 1 μg of total RNA using a Thermo Scientific RevertAid First-Strand cDNA synthesis kit (Thermo Scientific, USA) in a 20 µL reaction mixture containing 1 μL random hexamer primer, 2 μL deoxyribonucleotide triphosphate mix 10 mM, 4 μL of 5x reaction buffer, 1 μL ribonuclease (RNase) inhibitor, 200-unit RevertAid MMuLV reverse transcriptase enzyme, and appropriate RNase/deoxyribonuclease free water. The mixture was then incubated at 25°C for 5 minutes, 42°C for 1 hour, and 70°C for 5 minutes.
3.5. Quantitative Real-time Polymerase Chain Reaction
A real-time PCR was performed using a Thermo Scientific Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific, USA) reagent in an ABI Step-One Real-time PCR platform (ABI system, USA) with the following primers: (1) β-actin, forward: CCT TCC TGG GCA TGG AGT CCT; reverse: TGG GTG CCA GGG CAG TGA T; (2) PD-L1, forward: CTA TGG TGG TGC CGA CTA CAA, reverse: CTG CTT GTC CAG ATG ACT TCG
Gal-9, forward: CAG TGC TCA GAG TTC CAC A; reverse: TGA GGC AGT GAG CTT CAC AC; (3) CD155, forward: GGA CGG CAA GAA TGT GAC, reverse: CCA GTT GTT ATC ATA GCC AGA G.
The PCR reactions were amplified at 95°C for 5 minutes as initial denaturation, followed by 40 cycles at 94°C for 30 seconds, 57°C (β-actin), 61°C (PD-L1), 59°C (Gal-9), and 59°C (CD155) for 30 seconds, and extension at 72°C for 30 seconds. The PCR amplicon sizes were 174, 159, 118, and 124 bp for β-actin, PD-L1, Gal-9, and CD155, respectively. Each run was completed with a melting curve analysis to confirm the specificity of the amplification curves and the absence of primer dimers. The relative messenger ribonucleic acid (mRNA) levels were determined by normalizing the target gene’s fluorescence data to the housekeeping gene β-actin (2–∆∆Ct).
3.6. Statistical Analysis
GraphPad Prism software (version 8.0) was used to conduct statistical analyses and create graphs. Quantitative data are expressed as mean±standard deviation. The Kolmogorov-Smirnov test was employed to determine the normality of the distribution of the obtained data, followed by one-way analysis of variance and Dunnett’s test for multiple comparisons. P-values of less than 0.05 were considered statistically significant.
4. Results
4.1. Prevention of Proliferation of MCF-7 Cells by AKT/mTOR and BTK Signaling Pathway Inhibitors
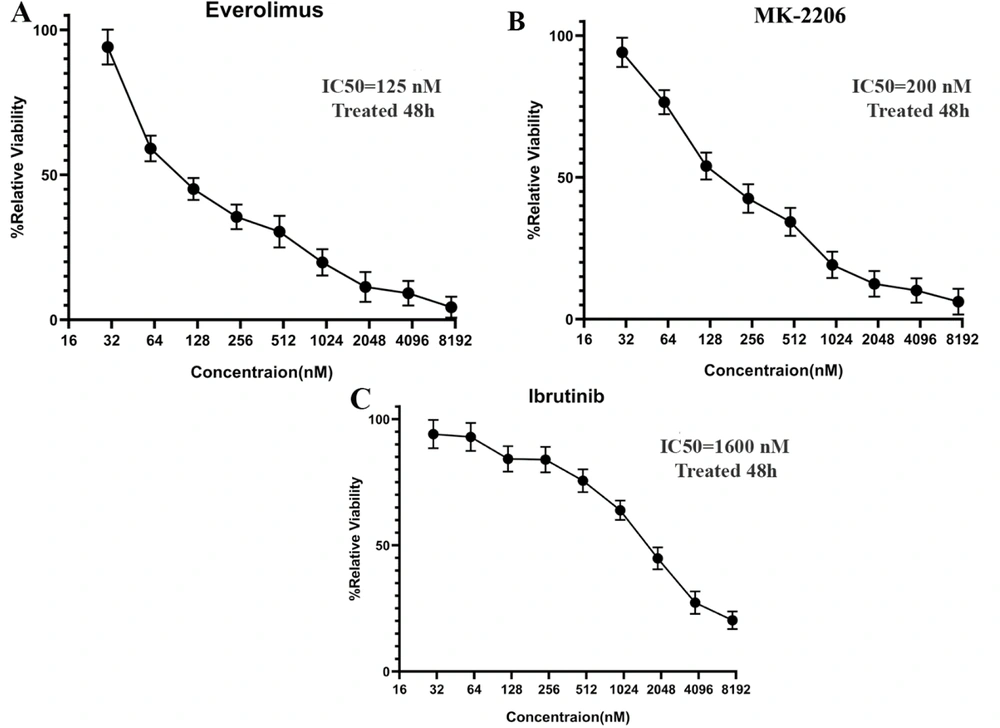
The effects of AKT/mTOR and BTK signaling pathway inhibitors on growth inhibition were surveyed in the MCF-7 cells. Initially, an MTT assay was used to determine the IC50 values of everolimus, MK-2206, and ibrutinib on MCF-7 cells after 48 hours of exposure to increasing concentrations from 16 to 8192 nM. Based on the IC50 results obtained from the MTT assay, MCF-7 proliferation was dose-dependently inhibited in comparison to the control group. The optimal doses of everolimus, MK-2206, and ibrutinib were observed to be 200, 320, and 2000 nM, respectively (Figure 1).
Half maximal inhibitory concentration (IC50) values of everolimus, MK-2206, and Ibrutinib on MCF-7 Cells. The MCF-7 cells (1 × 104 cells/well) were plated into 96-well culture plates and treated with increasing concentrations of A, everolimus; B, MK-2206; and C, ibrutinib for 48 hours. The data are shown as the mean ± standard deviation in triplicate. Untreated wells were used as controls.
4.2. mRNA Expression Profiles of Immune Checkpoint Ligands following Exposure to AKT/mTOR and BTK Inhibitors in MCF-7 Cells
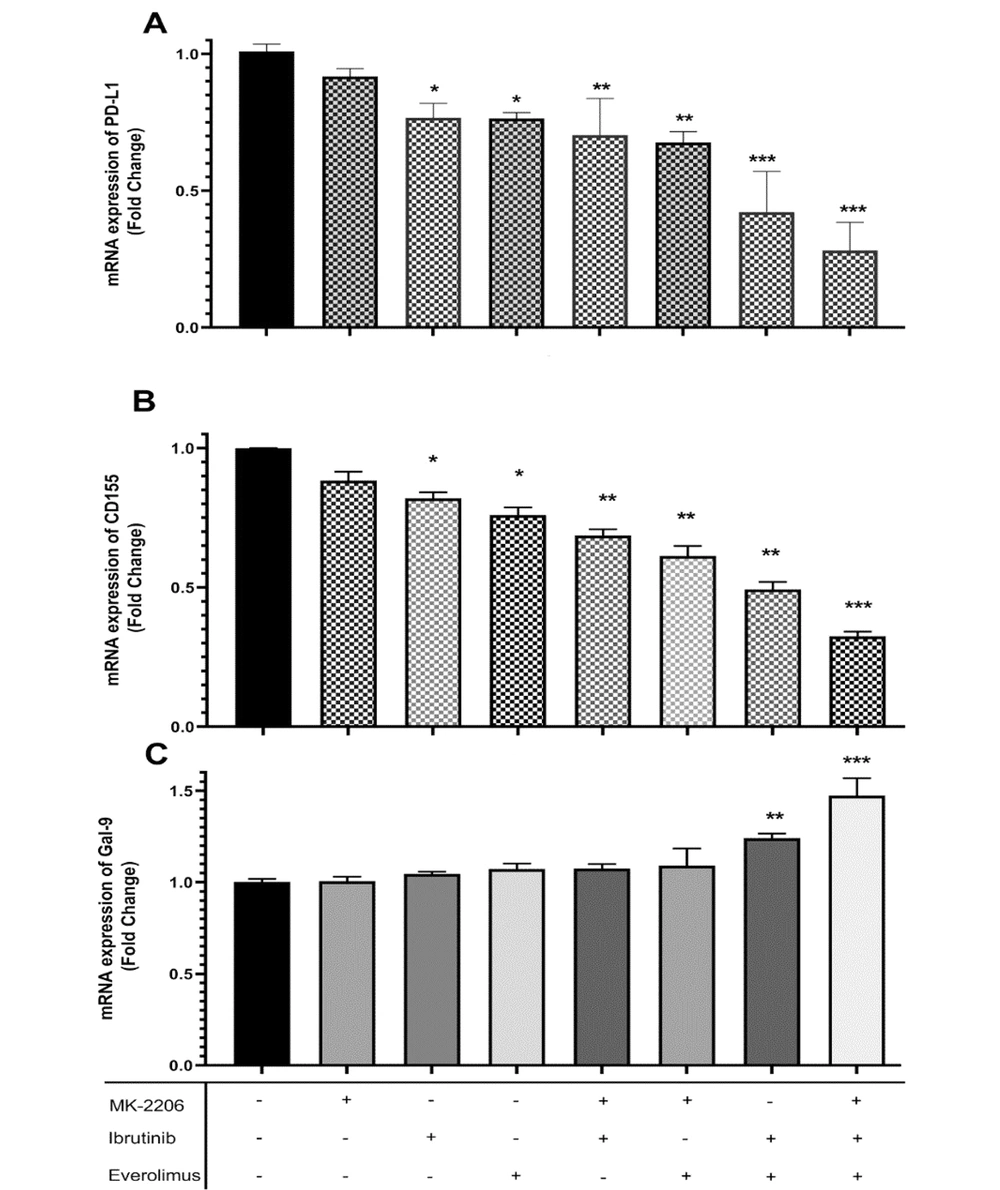
To better understand the molecular events underlying immune evasion in breast cancer cells, the effects of AKT/mTOR and BTK inhibitors were evaluated on the mRNA expression of immune checkpoint ligands PD-L1, CD155, and Gal-9 in MCF-7 cells. The obtained data of the present study demonstrated that PD-L1 and CD155 mRNA levels were significantly decreased in MCF-7 cells following a single treatment with everolimus and ibrutinib, but not with MK-2206 (Figure 2A and B). Intriguingly, PD-L1 and CD155 mRNA expression was significantly decreased following combination blockade with two or three drugs (P < 0.001 and P < 0.0001, respectively; Figure 2A and B). In contrast to the findings of PD-L1 and CD155, the results indicated no difference in the mRNA expression of Gal-9 between single-treated groups and controls. Cotreatment with everolimus and ibrutinib in the presence or absence of MK-2206 increased Gal-9 mRNA expression in cultured cells (P < 0.0001; Figure 2C).
A, Programmed death-ligand 1 (PD-L1); B, CD155; and C, galectin-9 (Gal-9) mRNA expression profile of MCF-7 cells treated with an optimized concentration of everolimus, MK-2206, and ibrutinib. Relative transcript levels (fold change) of the represented checkpoint ligands are represented as 2-ΔΔCt after normalization with β-actin as an internal control. Bars represent the mean ± standard deviation of three independent experiments (* P < 0.01, ** P < 0.001, *** P < 0.0001).
4.3. Protein Expression of Phosphorylated STAT3 following Exposure to AKT/mTOR and BTK Inhibitors in MCF-7 Cells
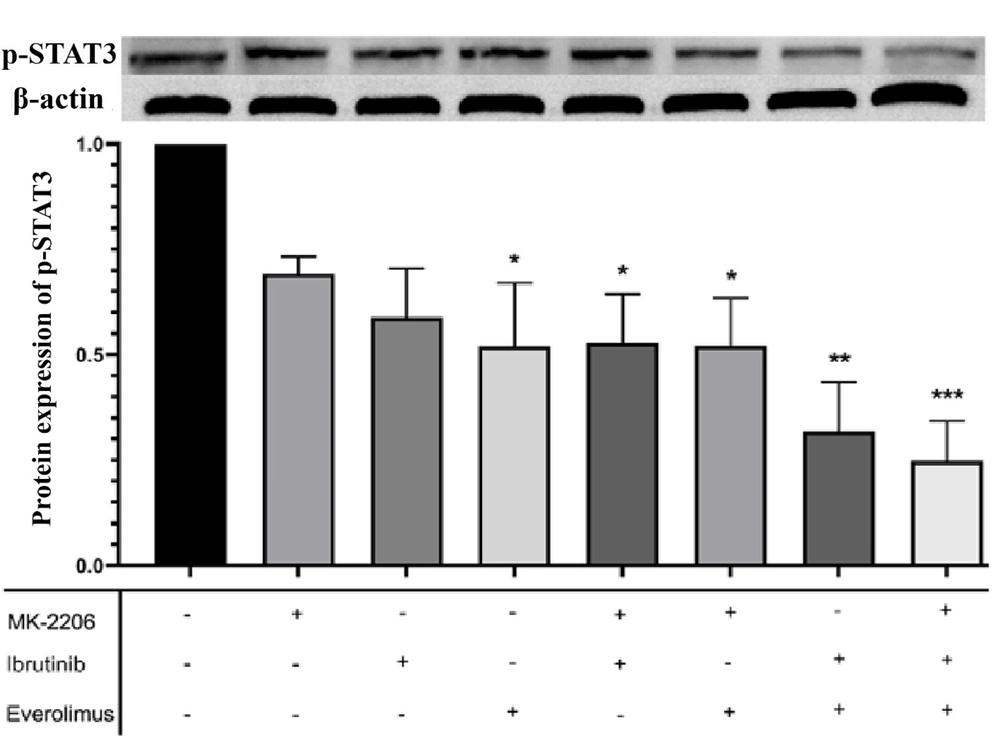
For the investigation of the intracellular signaling pathways involved in breast cancer cells’ immune evasion, this study used a western blot assay to determine the effects of AKT/mTOR and BTK inhibitors on the protein expression of p-STAT3 in MCF-7 cells. As illustrated in Figure 3, a single treatment with all three drugs resulted in a significant decrease in the level of Ser727-p-STAT3; however, only everolimus had a significant effect. As expected, the level of Ser727-p- STAT3 was decreased following combination treatment with two or three drugs (Figure 3).
Effects of everolimus, MK-2206, and ibrutinib on the phosphorylation of signal transducer and activator of transcription 3 (STAT3). Western blot analysis was applied to detect phosphorylated STAT3 (p-STAT3) expression in MCF-7 cells following treatment with the indicated small-molecule inhibitors. The expression levels of p-ser727-STAT3 were normalized to the levels of β-actin protein expression. Untreated cells were used as negative controls. Bars represent the mean ± standard deviation of three independent experiments (* P < 0.01, ** P < 0.001, *** P < 0.0001).
4.4. Correlation Analysis of Immune Checkpoint Ligand Expression and Phosphorylated STAT3 in MCF-7 Cells following Incubation with AKT/mTOR and BTK Inhibitors
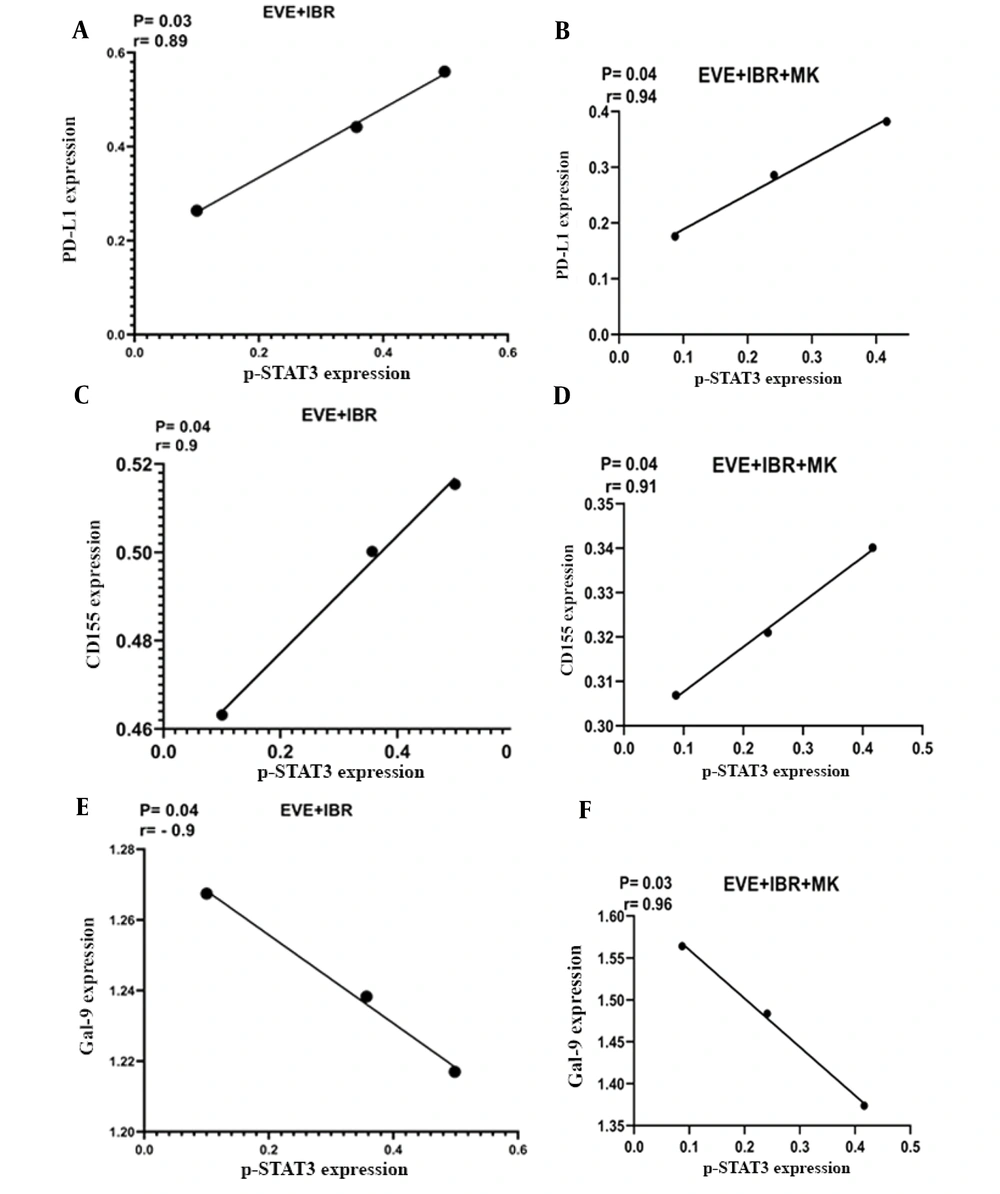
After measuring the mRNA expression of immune checkpoint ligands and the protein expression of p-STAT3, the obtained data were analyzed to find any correlations between the mRNA expression of PD-L1, CD155, and Gal-9 and the protein levels of p-STAT3 in MCF-7 cells treated with everolimus, MK-2206, and ibrutinib. The obtained data reflected a positive correlation between the expression of PD-L1 and CD155 in MCF-7 cells and the levels of p-STAT3 (Figure 4A - D). However, Gal-9 expression was negatively correlated with p-STAT3 protein expression (Figure 4E and F).
Correlation analysis of programmed death-ligand 1 (PD-L1), CD155, and galectin-9 (Gal-9) mRNA expression with protein expression of phosphorylated signal transducer and activator of transcription 3 (p-STAT3) in MCF-7 cells treated with everolimus, MK-2206, and ibrutinib. The PD-L1 and CD155 mRNA expression was positively correlated with p-STAT3 protein expression in MCF-7 cells treated by A, everolimus and ibrutinib; or B, everolimus, ibrutinib, and MK-2206. However, Gal-9 expression was negatively correlated with p-STAT3 expression in the same treatment (A and B). Real-time polymerase chain reaction and Western blot methods were performed to evaluate mRNA and protein expression, respectively.
5. Discussion
For the circumvention of the limitations of conventional cancer therapies, researchers are constantly searching for new molecular targets capable of selectively killing cancer cells. Small molecule agents and mAbs are considered the two major molecular targeting-related cancer therapy approaches currently used in clinical practice (26). Understanding the mechanisms underlying immune evasion mediated by tumor-intrinsic signals might help identify the new therapeutic strategies for optimizing cancer immunotherapy. The upregulation of immune checkpoint molecules on immune cells and their corresponding ligands on tumor cells is a critical mechanism for tumor cells to evade the host immune response (17, 20, 27, 28). These interactions result in the functional impairment of all immune cells, particularly T- and NK cell responses, ultimately allowing the tumor to escape the host immune system.
The present study examined the relationship between AKT/mTOR/BTK signaling pathways and the expression regulation of PD-L1, CD155, and Gal-9 immune checkpoint molecules following the treatment with three SMIs as mTOR, AKT, and BTK inhibitors. Previous studies have indicated that the AKT-mTOR pathway regulates PD-L1 expression in nonsmall cell lung cancer (15); the BTK pathway regulates PD-L1 expression in chronic lymphocytic leukemia patients via STAT3 (16); mitogen-activated protein kinase signaling pathways also regulate PD-L1 expression in lung adenocarcinoma (29). Mansour et al. demonstrated that the Notch and PI3K/AKT pathways promote PD-L1 expression in breast cancer stem cells (30). Several studies have shown that Raf-MEK-ERK-AP1 regulates CD155 expression in tumors via AP-2, nuclear respiratory factor, and sonic hedgehog (31, 32). Another study reported that ibrutinib inhibits cytokine production and PD-L1 expression resulting in decreased tumor vasculature density (33).
It has been recently demonstrated in human and mouse leukemia cells that the inhibition of intracytoplasmic signaling pathways by various SMIs interferes with the immune escape mechanisms of cancerous cells by the modulation of the immune checkpoint molecules (34, 35). This study showed that inhibiting mTOR and BTK, either alone or in combination, decreased the expression of PD-L1 and CD155 but did not affect their expression when an AKT inhibitor was used. This could be due to the compensatory signals produced by other tyrosine kinases, such as PI3k, mTOR, or BTK, when continuous AKT activation is inhibited. Concerning Gal-9, it has been demonstrated that the overexpression of this molecule reduces the spread of cancerous breast cells (36). Gal-9 can prevent metastasis by blocking CD44 attachment to hyaluronic acid. Another study found a positive correlation between increased Gal-9 expression and decreased metastasis in breast cancer patients (37).
The findings of the present study showed that the combined inhibition of mTOR and BTK pathways increased the level of Gal-9 mRNA expression. Therefore, the inhibition of mTOR and BTK pathways can reduce breast cancer metastasis through the upregulation of Gal-9. The simultaneous inhibition of mTOR, AKT, and BTK showed the same result, as expected. The aforementioned findings demonstrate that the inhibition of mTOR, AKT, or BTK alone or in combination exhibits various effects on immune escape mechanisms.
In this study, the antiproliferative activity of everolimus, MK-2206, and ibrutinib was determined by measuring their relative IC50 values. Compared to the 200 nM IC50 value obtained in the current study for everolimus in MCF-7 cells, Leung et al. reported IC50 values of > 100 nM for two sublines of MCF-7 cells, TamC3 and TamR3 (38). In another study, the IC50 values for everolimus were reported as > 50 nM in low glucose and 29 nM in high glucose conditions in MCF-7 cells (39). Although the calculated IC50 value for ibrutinib was obtained at 2000 nM in the present study, the IC50 value of ibrutinib was indicated at 6400 nM in MCF-7 cells in another study (40). Surprisingly, an IC50 of 30 nM has also been reported for ibrutinib in the MCF-7 cell line, which is much lower than those of the present study and the aforementioned study (41). Regarding MK-2206, previous studies on MCF-7 and BRCA1-deficient breast cancer cells reported IC50 values of < 500 nM and 1250 nM, respectively (42, 43). Similarly, the IC50 of MK-2206 in the current study was obtained at 320 nM. The observed variations in the IC50 values in different studies can be related to the various types of assays and conditions leading to diverse results.
The STAT3 was identified as a critical mediator of cancer SMI therapy for modulating immune checkpoint molecules via these cancer-related signaling pathways. This study examined STAT3 autophosphorylation to determine whether STAT3 activation is associated with PD-L1, CD155, and Gal-9 transcription. The results indicated a significant decrease in p-STAT3 expression in MCF-7 cells treated with everolimus and ibrutinib alone or in combination, which was positively correlated with the expression of PD-L1 and CD155, but inversely associated with Gal-9 expression. Furthermore, other studies demonstrated that STAT3 regulates PD-L1 expression in tumor cells, allowing them to evade immune surveillance (44-46). Moreover, ibrutinib was shown to inhibit the STAT3 pathway in human glioblastoma (47). The aforementioned findings emphasize the critical role of the STAT3 transcription factor in the expression regulation of checkpoint ligands and the immune evasion of breast cancer cells.
In addition, previous research has shown that the sustained activation of STAT3 in the nucleus directly modulates the actions of nuclear factor kappa B (NF-κB). When STAT3 is phosphorylated at Tyr705 in the nucleus, the connection between STAT3 and NF-κB is strengthened (48). The STAT3/NF-κB signaling pathway is involved in tumor angiogenesis and invasiveness, and both NF-κB and STAT3 have been implicated in cancer cell invasion, metastasis, angiogenesis, and immune evasion (49). Ibrutinib was reported to significantly inhibit Janus kinase/STAT signaling and mitigate the effects of CpG stimulation on NF-κB signaling (50). Additionally, ibrutinib enhances the efficacy of bortezomib in primary tissues by inhibiting the BTK/NF-κB p65 signaling axis (51). On the other hand, BP-1-102, a STAT3 inhibitor, was shown to inhibit NF-κB activity, probably by influencing the crosstalk between STAT3 and NF-κB (52). Notably, it has been demonstrated that NF-κB regulates PD-L1 expression in cancer cells, which was associated with strong crosstalk between STAT3 and NF-κB (53, 54). In this regard, the effects of applied SMIs on STAT3 expression in the current study might inhibit NF-kB activity, thereby decreasing the expression of immune checkpoint ligands.
5.1. Conclusion
Overall, AKT/mTOR and BTK signaling pathways are not only involved in cancer cell proliferation, tumorigenesis, and clinical progression but also regulate the escape of tumors from immunological surveillance and anticancer immune responses. Since AKT/mTOR and BTK pathways regulate immune checkpoint expression, the pharmacological inhibition of these cascades in cancers will be more advantageous due to the inhibition of tumor growth and blockade of immune evasion activity mediated by intrinsic signals. On the other hand, because AKT/mTOR, BTK, and other signaling pathways are required for normal antitumor immunity, the therapeutic inhibition of these pathways should be as selective as possible in targeting cancer cells while having minimal adverse effects on the host immune system. Combinational therapy approaches that interfere with immune escape mechanisms might represent a novel and promising therapeutic strategy for patients with breast cancer.