1. Context
The foremost pneumonia incident was registered in Wuhan city, China, in December 2019. Later, this novel virus was identified as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which robustly spread globally. On 11 March 2020, the World Health Organization (WHO) declared this virus a mostly infectious malady (1-3). Globally, 286,904,582 total active cases of COVID-19 involving 5,446,652 deaths were reported to WHO on 28 December 2021. Recently, the emergence of the deadly novel coronavirus variant, i.e., the Omicron variant (B.1.1.529), has set flames globally with its robust outbreak without any signs and symptoms (4). The Omicron coronavirus variant was first acknowledged on 24 November 2021 in Botswana, South Africa, and later named the variant of concern (VOC) on 26 November 2021. It has been noted that omicron showed a considerable number of mutations, approximately 32 in the spike protein region. However, the delta variant revealed 16 mutations in the spike protein region. Today, Omicron has been considered more infectious than the original coronavirus, particularly the delta variant.
Before the mitigation of corona variants such as alpha, beta, delta, and omicron came into existence, the death toll was augmenting every day, which is regarded as a significant global problem (5). Moreover, it is challenging for the world to prevent COVID-19 by discovering an ideal drug/vaccine. Traditional medicine has been followed for ages. Nevertheless, plant-related products such as Garlic (Allium sativum L.) depicted in (Figure 1) have been regarded as significant basil for over 4000 years, and cheers to its diversity plus abundant medicinal value embraced as a regular seasoning among household meals (6, 7).
Herbal plant-based items are tremendous initial points for a breakthrough in drugs. Traditional remedial extracts from plant-related compounds have been practiced since ancient times in curating various diseases, including viral infections (8, 9). It has been reported in the literature on inhibitory actions of replications among multiple viruses such as SARS, herpes simplex virus 2 (HSV-2), human immunodeficiency virus-1 (HIV-1), and hepatitis B virus (HBV) (10). However, based on several literature descriptions of floral medicinal abundance, it has been recommended that herbal extracts possess a broad spectrum of actions against drug-resistant viruses, which might be associated with their versatile constituents (11). The mode of action of these extracts and their purified components displays antiviral activity based on virus strains and their life cycle from attachment, synthesis, replication, and assembly (12).
Nonetheless, herbal products possess immunomodulatory properties to fight viral infections. Therefore, our review article primarily focused on the development of herbal medicines as antiviral agents, which has the potential to boost immunity and provide antiviral defense (13). Amongst herbal plants, A. sativum L. (garlic) is a bulbous basil from the Alliaceae family indigenous to Central South Asia. Garlic is an annual basil specifically exploited for gastronomic and other ethereal perseverance for many years. From the ancient epoch, garlic has been routinely utilized as a functional food, flavor, and seasoning herb, with previous reports on medicinal herbs displaying effectiveness against viral infections. Our prime focus is to discuss garlic's mode of action, which embraces several components like allyl methyl thiosulfinate, allicin, methyl allyl thiosulfinate, and ajoene. Also, interaction with the SARS-CoV-2 receptor displays a prospective approach for treating the malady. In forthcoming years, it can develop as a possible source of antiviral drugs for curtailing the outburst of COVID-19.
2. Taxonomic Classification, Unani Description, and Common Names
Allium actually belongs to the onion genus, such as Allium cepa (onion), A. ascalonicum (shallots), A. porrum (leeks), and A. schoenoprasum (chives). In addition, Allium sativum is further classified into two subspecies, A. sativumvar. sativum acknowledged as soft neck garlic, and the other one, Allium sativumvar. ophioscorodon, recognized as hard neck garlic. Nonetheless, both have an underground bulb made up of cloves; prophylls are encircled by dry membranous skins held together by a basal plate. Eventually, garlic contains 6 to 11 cloves with a scape at the top, giving rise to bulbils (Table 1 and Figure 2).
| Variables | Values |
|---|---|
| Kingdom | Plantae |
| Subkingdom | Tracheobionta |
| Clade | Angiosperms |
| Clade | Monocots |
| Super-division | Spermatophyta |
| Order | Lillales |
| Family | Alliaceae |
| Subfamily | Allioideae |
| Class | Monocotyledons |
| Sub-class | Liliidae |
| Genus | Allium |
| Species | Sativum |
2.1. Unani Categorization
- Botanical name: Allium sativum. L
- Unani name: Lehsun, Fum, Thoum, Thum
- Properties: Mizaj 3rd order warm and dry
- Synonyms: Allium sativum, Garlic, Saun Taum, Lehsun, Lashan, lahsun.
- Muzir: Mehrooreen (for persons with hot temperament)
- Maza: Bitter and acrid
- Mukhrij: Expels Balgham (phlegm)
- Boo: Radio-active and acrid
- Nafa-e-Khas: Phlegmatic and stomach-related diseases
2.2. Familiar Names
- Synonyms: Allium sativum L.
- Sanskrit: Lasuna, Rosona, Yovanesta
- English: Garlic, poor man's treacle
- Arabic: Saun Taum
- German: Knoblauch, Lauch
- Greek: Allidion, Skorodon
- Italian: Aglio
- Chinese: Syun tauh
- Urdu: Lehsun
- Malayalam: Veluthulli.
- Bangali: Rosun
- Hindi: Lashan, lahsun
3. Viral Structural Proteins and Promising Herbal Drugs as Therapeutic Agents
The structural protein viral RNA encodes for envelope (E), membrane (M), nucleocapsid (N), and spike (S) proteins (13-16). Moreover, soluble protein (N) encloses nucleocapsid packages of the RNA genome. The small E protein is encountered with approximately 76 - 109 amino acids encompassing the predicted hydrophobic domain that is indispensable for association with the main membrane M-protein in the virion envelope (17, 18). However, the core route entry for the virus is an addon with protein S, which attaches to the host cells receptor, i.e., angiotensin-converting enzyme 2 (ACE2). The plasma membrane endocytoses the coronavirus and instigates its entry into the host cell cytoplasm. Later, it releases the RNA genome to be translated into structural proteins (N, M, E, and S) (19). With the effect of its robust transmission of infectivity, scientists are attempting to find novel drugs for combating COVID-19 (20). Hence, repurposing drugs revealed promising targets for the near future in improving therapeutic agents/vaccines against deadly human coronavirus disease (21).
4. Docking Simulation Studies
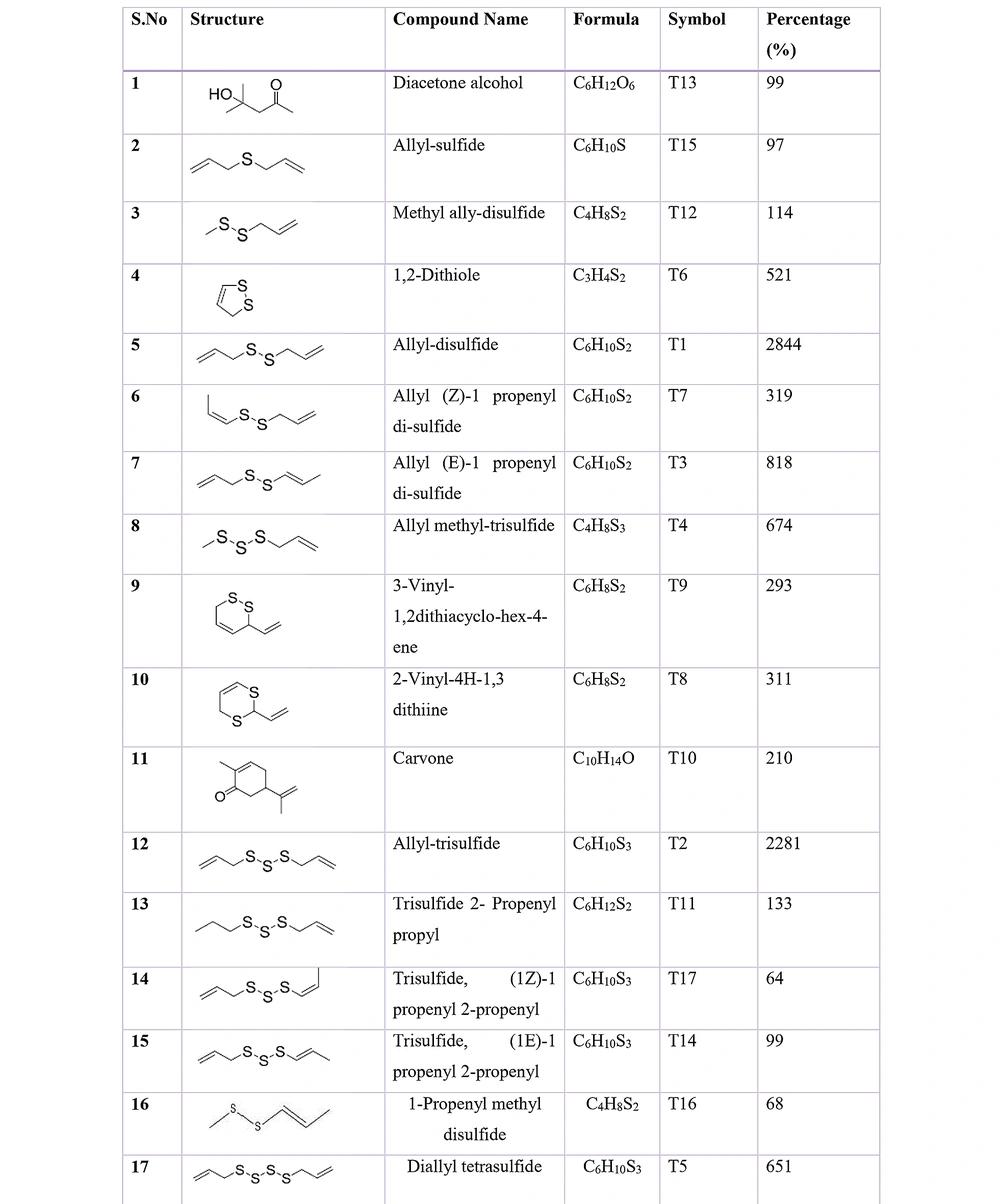
Many scientists have recognized the structure of the main protease of SARS-CoV-2. It has a serine-type Mpro (chymotrypsin-like protease (3CLpro) protease with amino acids like Thr24, Thr26, and Asn119) in the active site portion. Nonetheless, this protease is primarily accountable for one to two kinds of SARS-CoV and consequently displayed that protease is responsible for the proteolytic maturation of SARS-CoV-2 (22). However, previous literature reported the antiviral actions of several herbal extracts against coronavirus protein targets (23). Amongst in-silico methods, garlic has revealed noticeably inhibitory activities against the coronavirus. The compounds of A. sativum are alliin, S-(allyl/ methyl/ ethyl/propyl)-cysteine, S-propyl L-cysteine, and S-allymercapto-cysteine, which was regarded as potent bioactive agents to inhibit the Mpro of SARS-CoV-2 in H-bonds with support of protease. Several literature reports on molecular docking analysis depicted that alliin, among other Organosulfur Compounds (OSCs), has a higher antiviral capability to mitigate COVID-19 (Figure 3 and Table 2). Moreover, much research must be carried over to examine whether this bioactive component alone can provide efficacy or needs a combination approach to completely eradicate SARS-CoV-2 with very few side effects and toxicity (24).
| Name of Compound | Compound-Protein Interaction | Docking Score Energy (kcal/mol) |
|---|---|---|
| Allyl-disulfide | T-1 with ACE-2 receptor | -12.83 |
| Allyl- trisulfide | T-2 with ACE-2 receptor | -12.75 |
| Allyl (E)-1-propenyl-disulfide | T-3 with ACE-2 receptor | -9.07 |
| Allyl methyl-trisulfide | T-4 with ACE-2 receptor | -12.49 |
| Diallyl-tetrasulfide | T-5 with ACE-2 receptor | -14.05 |
| 1,2 di-thiole | T-6 with ACE-2 receptor | -7.89 |
| allyl (Z)-1-propenyl-disulfide | T-7 with ACE-2 receptor | -9.04 |
| 2-vinyl-4H-1,3-dithiine | T-8 with ACE-2 receptor | -11.83 |
| 3-vinyl-1,2-dithiacyclohex-4-ene | T-9 with ACE-2 receptor | -10.5 |
| Carvone | T-10 with ACE-2 receptor | -8.50 |
| trisulfide, 2-propenyl propyl | T-11 with ACE-2 receptor | -14.00 |
| methyl allyl disulfide | T-12 with ACE-2 receptor | -10.30 |
| Diacetone alcohol | T-13 with ACE-2 receptor | -9.70 |
| trisulfide, (1E)-1-propenyl 2-propenyl | T-14 with ACE-2 receptor | -9.50 |
| Allyl-sulfide | T-15 with ACE-2 receptor | -9.38 |
| 1-propenyl methyl-disulfide | T-16 with ACE-2 receptor | -8.06 |
| trisulfide, (1Z)-1-propenyl 2-propenyl | T-17 with ACE-2 receptor | -8.06 |
5. Mechanism of Antiviral Agents from Allium sativum. L
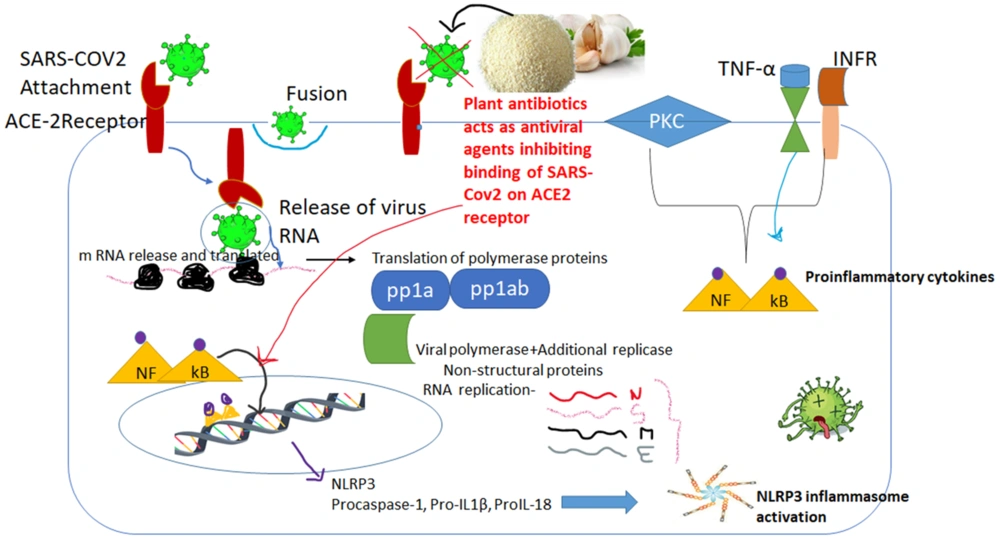
Like other beta-coronaviruses, SARS-CoV-2 modifies to bind to the host cell receptor ACE-2 and affects the alveolar lungs (25, 26). Upon binding to the receptor, SARS-CoV recruits the protease enzyme for cleaving the S protein into S1 and S2 domains, resulting in endocytosis of virion inside the host cell. The viral RNA genome translates its protein by activation through NF-κB alleyway, resulting in transcription of the pro-IL-1B gene and NLRP3 inflammasome formation (27). The primary role of NLRP3 inflammasome is to exert antiviral immune responses inside the host. Several literature reports have shown that activation of NLRP3 inflammasome in IAV and West Nile virus (WNV) (28) inhibits viral replication. However, the mouse model study also depicted the actions of NLRP3 inflammasome in inhibiting viral replication and mitigating mortality among animal models (29). The primary step is TNFR activation, which triggers NF-kB and further promotes the expression of NLRP3, pro-IL-1b, and pro-IL-18 (30). Nonetheless, the assemblage of the NLRP3 inflammasome instigates the auto-cleavage of procaspase-1 (31, 32), mediates proteolytic processing of pro-IL-1b and pro-IL-18, and induces viral cell death known as pyroptosis (33). Thus, NLRP3 inflammasome instigation acts as a spear/double-edged sword, as it has the potential to act as a defensive agent against viral infection (Figure 4).
Chemical constituents present in Allium sativum inhibit the binding of SARS-CoV-2 to the ACE2 receptor. In general, if the virus binds, it triggers a cellular cascade from NF-kB, which activates AKT/PKB and further promotes Pro-IF1βand Pro-IF18, resulting in NLRP3 expression and exerting antiviral immune responses in the host. The binding alliin compound, in turn, activates the inflammasome and results in viral protein degradation.
5.1. Repurposed Compounds Act as New Therapeutic Agents Against COVID-19
Table 3 displays the purpose of repurposed drugs and their confirmed preclinical studies. The table shows several repurposed compounds used against various other diseases revealed to possess therapeutic potential even against SARS-CoV-2. Mainly repurposed compounds targeting chief corona protease were selected. The selected compounds are based on their complementarity between ligand-receptor dockings, which displayed root mean square deviation (RMSD) and helped identify the higher affinity compounds. Later, the compounds were exposed to molecular dynamic simulations, which show their ADMET properties, i.e., absorption, distribution, metabolism, and excretion, studied with AdmetSAR and Swiss ADME software (34).
Nonetheless, Kim et al. (35) employed two computational methods for recognizing subsisting drugs that are FDA-approved to prevent or mitigate clinical infections of respiratory distress. The initial methodology predicted the artificial intelligence-based binding affinity platform for identifying suitable medicines that could prevent the entry of infectious viral cells inside the host. The entrance of the virus is mainly through an addon with or transmembrane serine protease 2 (TMPRSS2) along with angiotensin-converting enzyme 2 (ACE2). The second method employed was the disease cancelling technology (DCT) manifesto to identify compounds that could attenuate the expression of genes instigated by coronaviruses (36).
The development of artificial intelligence technology in pattern recognition through deep learning and machine learning program displayed several drugs, which are FDA-approved pharmaceuticals and nutraceuticals for targeting coronavirus envelope (E) protein mainly related to the release of the virus inside the host cell. Hence, diverse computational methods are used to predict repurposed drugs' efficacy against the current pandemic.
| Drugs | Summary of Drug Actions with Confirmatory In-Vitro Clinical Data | References |
|---|---|---|
| Baricitinib | An artificial intelligence (AI) platform recognized baricitinib agents as a probable COVID-19 therapy. However, detailed information about its mode of action is limited. | (37, 38) |
| Remdesivir; aprepitant; valrubicin; fulvestrant | With the support of the artificial intelligence approach, we can select particular repurposed drugs, selectively directing the central coronavirus protease (6LU7) utilizing receptor-ligand interactions with liquid simulation algorithms to recognize greater binding compounds. In addition, drug compounds, after molecular dynamic simulations, are directed for absorption, digestion, metabolism, excretion, and toxicity studies. | (34) |
| Cinametic acid; guaifenesin; lauric acid; ascorbyl palmitrate; nabumetone; octacosanol; palmidrol; salmeterol; nafcillin | Advancement in AI machinery with much machine knowledge and deep learning program was employed for comprehension of in-silico methodologies along with pattern identification technology for monitoring FDA-approved pharmaceuticals and nutraceuticals to target the CoV envelope (E) protein. Protein E is indulged in the association and discharge of the virion interior into the host. A multitude of open-source drug databases was considered, such as Enamine Bio-reference compounds (https://www.enaminestore.com/products/bioreference-compounds, accessed on 24t February 2022) | (39) |
| Brequinar; clofazimine; tolcapone; bedaquiline; conivaptan; gemcitabine; vismodegib; celecoxib | Earlier, artificial intelligence technology was established to recognize repurposed/ traditional drugs with anti-coronavirus activities. With the introduction of machine learning programs, further prediction of medicines with the help of AI machinery was later tested for their in-vitro potential against a feline coronavirus. However, the deep neural network algorithm is currently employed to recognize the most applicable descriptors for the fabrication of AI prediction models. | (40) |
| Lisinopril; spirapril; captopril | Artificial intelligence-amalgamated automatic modeling platform BIOiSIM was employed to simulate the systemic treatment of blockers of calcium channels and ACE compounds in tissues related to the COVID-19 pathogenesis, namely the disposition and site-of-action penetration (in-silico modeling). The most scalable dynamic computer-based prediction is BIOiSIM, which predicts in-vivo pharmacokinetic-pharmacodynamic (PK-PD) phenomena. | (41) |
| Albuterol; artemisinin; amprenavir; chloroquine; ciprofloxacin; fluoroquinolones; hydroxymethylglutaryl-coa reductase inhibitors; cyclosporine; quinolone; antibacterial agents; methotrexate; zidovudine; suramin | To predict the link, an AI text-mining model was developed. Various other approaches, such as SemNe, and biomedical knowledge graph, were employed to predict missing links in biomedical literature, which was helpful in drug repurposing. However, RotatE, TransE, and CompleX methods are based on the knowledge of graph embeddings and outcomes predicted with web application | (42) |
| Fulvestrant; aprepitant; valrubicin; remdesivir | For repurposed drugs, a machine-learning approach was utilized. Specifically, repurposed drugs targeted the main coronavirus protease (6LU7) employing ligand-receptor docking, which was optimized using liquid simulation algorithms to recognize high-affinity compounds. In addition, drug candidates were subjected to molecular dynamic simulations and further examined for adsorption, distribution, metabolism excretion, and toxicity studies (ADMET). | (34) |
| Dihydroergotamine; venetoclax; conivaptan; eltrombopag; ergotamine; idarubicin; ledipasvir; lifitegrast; lumacaftor; eribulin; nilotinib; nystatin; paritaprevir; ponatinib; regorafenib; rifapentine; simeprevir; teniposide; trabectedin; trypan blue; ivermectin; velpatasvir | Different in-silico approaches were combined for decreeing virtual drugs or utilizing molecular docking and supervised machine learning algorithms. However, all were used in a determined manner to recognize repurposed drug activity against COVID-19. | (43) |
6. Docking Methodology for Studying Targeting on Mpro
A virtual screening approach was used to study the substrate binding precisely. Nonetheless, the deep docking approach in association with Glide was utilized for investigating the inhibitory action of approximately 1.3 billion compounds from the ZINC15 library and among 1,000 recognized potential compounds which displayed higher scores of docking onto the active site of SARS-CoV-2 Mpro (44). With the support of the computational search for Mpro inhibitors, various screening steps are involved, such as SMINA docking, shape screen, clustering, Glide docking, and pharmacokinetic descriptors. Further usage of MD simulations can assess approximately higher than 606 million compounds from the ZINC database for toxicity assessment (45). However, previous reports recognized the multitarget approach, such as targeting nine natural molecules to Mpro (46). In other reports, docking analysis has revealed not only targeting of Mpro but additional sites such as ACE2 and RdRp. Our study depicted allicin binding to the ACE2 receptor and its docking outcomes. Nonetheless, several researchers have identified and shown promising binding capacity that must be screened further, and clinical trials need to be conducted (47-50). Experimental observation of all the discovered compounds is noteworthy to provide potent drugs against COVID-19.
7. Plant-Derived Products Have Abundant Secondary Metabolites Which Act as Potent Antiviral Agents
From the primeval period, Indian herbs have been used for therapy and defensive approaches against various maladies, including SARS-CoV-2. The primary purpose of using these herbs primarily boost immunity as they possess immunomodulatory properties. The Indian plants have preventive and prophylactic actions encouraging utilization for various maladies (51). However, regular garlic consumption in our diet increases immunity and protects us as a defensive agent against different pathogenic organisms (52). Previous literature reports confirmed that medicinal plants possess secondary metabolites which can treat several disease states and were established to terminate avian influenzas virus (H9N2) (53).
Several reports have supported that garlic (A. sativum) is recommended amongst various medicinal herbs as a commonly utilized home remedy, specifically during this pandemic. Garlic is a functional food used in conventional medicine from ancient times to treat several diseases and can act as a better candidate due to its antimicrobial, antioxidant, anti-inflammatory, anti-thrombotic, immunomodulatory, and antiviral activities (54). The primary bioactive metabolites of garlic are divided into two groups: (1) sulfur-containing; and (2) non-sulfur-containing compounds. Alliin and allicin belong to chief sulfur-comprising compounds, although the chief sulfur-free bioactive compounds comprise saponins and flavonoids (55). Garlic has the potential to curb SARS-CoV-2, which can be examined in the in-silico studies by the configuration of hydrogen bonds amid amino acids that occurs at the addon site of the main structural protease of SARS-CoV-2.
A few reports also revealed that garlic could reduce COVID-19 among affected patients and that adding garlic-rich food daily significantly enhanced T helper cells, cytotoxic T cells, and NK cells. Furthermore, there are reports on the downregulation of leptin, leptin receptor, TNF-α, IL-6, and proliferator-activated receptor gamma (PPAR-γ) (56). Therefore, garlic can be regarded as one possible choice for the prevention of COVID-19 since it has the potential to modulate cytokine secretion, phagocytosis, macrophage activation, and immunoglobulin production (57, 58).
8. Beneficial Effects of Conventional Dietary Food Against Repurposed Drugs
The multistep drug development process usually takes more than five years to ensure the new chemical's safety and effectiveness. Antiviral medications targeting coronaviruses employ several approaches, such as target-based drug development, extensive phenotypic screening of chemical libraries, and empirical testing of well-established antiviral medications. Only remdesivir and a few neutralizing antibodies have been licensed by the US FDA for treating COVID-19, even though an increasing number of drugs have been demonstrated to have anti-coronavirus activity in vitro and in vivo. New antiviral medications are desperately needed, as remdesivir's therapeutic results remain debatable. Here, we highlight the creation of novel SARS-CoV-2 infection therapies that have been carried out in fundamental research, given the urgency of the SARS-CoV-2 outbreak (20, 59-61).
9. Conclusions
The current outbreak of SARS-CoV-2 is regarded as a threat to the human populace since the non-availability of proper vaccines/drugs for its therapy. Our paper mainly focuses on traditional herbs regularly used as spices in food preparation, now acknowledged to revamp immunity against several viral infections, including the coronavirus. Although several traditional herbs have been studied, to our knowledge, garlic contains an allicin compound that can work against virus entry into the host. Further research must be conducted to investigate their effectiveness, safety, and clinical efficiency. Nonetheless, we highlighted the repurposed drugs, their clinical significance, herbal drugs, and their formulations with a broad spectrum of antiviral actions. The whole universe is facing the pandemic situation, and our paper highlighted the significance of garlic and provided a path for researchers to concentrate on clinical trials to release the compound, which could act as a novel vaccine in the rapid curation of the current pandemic.