1. Background
Fetal abortion is one of the critical and controversial issues in scientific, social, and academic contexts (1). Abortion refers to the termination of pregnancy before the 20th week or the remission of the fetus weighing less than 522 grams (2). Abortion may occur spontaneously or through induction. An abortion that occurs spontaneously is also known as a miscarriage, but induced abortion occurs when intentional measures are taken to end a pregnancy (3). For determining the reason for abortion, laboratory tests have been divided into six broad groups: genetic, anatomic, metabolic, endocrinology, immunologic, and microbiologic (4). In each of the mentioned categories, different methods are applied.
A post-mortem examination consists of a methodical examination of a corpse to find the cause, mode, and manner of death (5). Among various aspects of post-mortem examination containing pathology, histology, bacteriology, etc., post-mortem bacteriology has been applied in multiple studies, not only for distinguishing the presence of an underlying infectious disease but also for determining the cause of death (6, 7). Heart blood, cerebrospinal fluid (CSF), and spleen tissue are the most common specimens for post-mortem microbiological cultures (8). Blood culture is the most used specimen among these common specimens in various research studies. Tissue and fluid samples must be taken 1 - 15 hours after death; in this way, the possibility of post-mortem bacterial invasion will be diminished. Various species of bacteria have been mentioned to be associated with abortion, but there is no evidence of a unique organism causing abortion (5). However, we examined the bacteriological heart-blood samples of aborted fetuses to determine the bacterial profile involved in their abortion. A post-mortem exam is one of the choices that can be used as a practical examination to determine what has caused the abortion (9).
2. Objectives
Given the point of view that post-mortem bacteriologic studies can be helpful and essential to determine the reason for an abortion, the current study was conducted as the first study in Shiraz, southwest Iran, to find out the profile of the involved bacteria and their antibiotic susceptibility pattern to design and implement probable appropriate interventions.
3. Methods
3.1. Sample Collection and Study Design
This cross-sectional study was carried out from 2016 to 2017 at the medical laboratory of Hazrat Zeinab, a major referral hospital for obstetrics and gynecology teaching in Shiraz, southwest Iran. As the following description, 153 blood samples were gathered from the heart of aborted fetuses. The samples were tested as previously described by Carter (10). Briefly, a 5-cc syringe was delivered to the microbiology laboratory 1 - 15 hours after birth. Then, the blood sample was inoculated into the Tryptone soy broth (TSB) (HiMedia, India) bottle, incubated at 37°C, and examined daily for turbidity and growth. In the case of observing turbidity in the TSB bottle, after 24 h, a sub-culture was done onto the blood agar (BA) plate (HiMedia, India), chocolate agar plate (CA) (HiMedia, India), and eosin-methylene-blue agar (EMB) (HiMedia, India). If no turbidity was observed after 24 h, at intervals of 48 h, 72 h, and 10 days after the first culture subculturing in plates of BA, CA, and EMB agar was done.
3.2. Biochemical Standard Tests
Bacterial identification at the species level has been accomplished according to the biochemical standard tests as explained by Shoaib et al. (11). Briefly, the isolates have been divided regarding Gram staining results; the species of Gram-negative (GN) rod isolates were detected as illustrated in Table 1. Furthermore, Gram-positive (GP) cocci isolates have been identified as Staphylococcus spp. and Streptococcus spp. Using morphology (cluster and pairs or chains), catalase (+ and -), oxidase (- and -), and hemolysin (β and β), respectively. Eventually, the coagulase test for Staphylococcus spp. has been performed.
| Bacterial Species | Biochemical Tests | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-Stain | Catalase | Oxidase | TSI | H2S | Indole | MR Test | VP Test | Citrate | Urease | Motility | |
| Acinetobacter spp. | - | + | - | Alk/Alk | - | - | - | - | + | V | - |
| Escherichia coli | - | - | - | A/A | + | + | + | - | - | + | |
| Klebsiella spp. | - | - | - | Alk/A | - | - | - | + | - | + | - |
| Enterobacter spp. | - | - | - | A/A | - | - | - | + | + | - | + |
| Pseudomonas spp. | - | - | - | Alk/Alk | - | - | - | - | + | + | + |
Abbreviations: Alk, alkaline; A, acid; TSI, triple sugar iron agar; MR, methyl red; VP, Voges Proskauer; V, variable.
3.3. Antimicrobial Susceptibility Testing
The isolated bacteria were further tested to demonstrate the antimicrobial susceptibility pattern (ASP) using the Kirby-Bauer disc diffusion method as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2018) (12). The ten tried antibiotic discs were as follows: Cefoxitin (30 µg), cefixime (30 µg), cefepime (30 µg), ampicillin-Sulbactam (10/10 µg), imipenem (10 µg), gentamicin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), tetracycline (30 µg), and trimethoprim/sulfamethoxazole (1.25/23.75 µg) (Mast, UK). The Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were tested for quality control.
4. Results
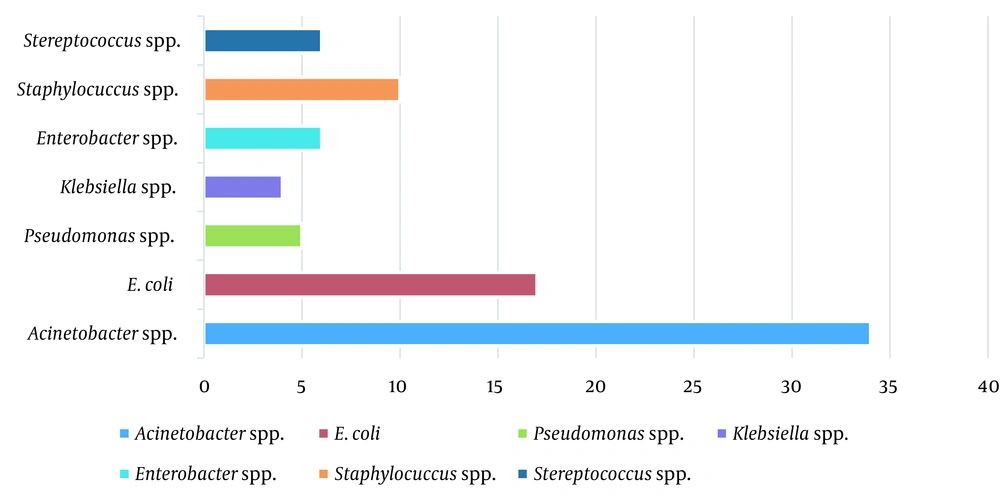
Out of 153 samples, 82 (53.6%) yielded positive results, among which 66 isolates (80.5%) were GN and 16 (19.5%) were GP cocci. In GN isolates, Acinetobacter spp. was the most frequent bacterium with 51.5% (34/66 isolates), and Staphylococcus spp. with a frequency of 62.5% (10/16 isolates) was the most frequent GP one. The bacterial profile of isolated strains from the heart-blood samples of septic abortion isolates is shown in Figure 1.
4.1. Antimicrobial Susceptibility Testing
Generally, the highest resistance rate among GN isolates was observed against cefixime (68.2%), followed by amikacin (63.6%), gentamicin (63.6%), and ciprofloxacin (56.1%). Also, among GP isolates, the highest resistance was against ciprofloxacin (62.5%), and the most effective one was tetracycline, for which no case of resistance was detected. For Staphylococci spp., the most frequent isolate, the highest resistance rate was against ciprofloxacin and cefoxitin (100%), but all isolates were susceptible to cefepime and tetracycline. Two detected non-hemolytic Streptococci spp. were sensitive to all the tested antibiotics (Table 2).
| Isolated Bacterial Species | The Antibiotics’ Tested Name | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FOX | CFM | FEP | SAM | IMP | GEN | AMK | TET | CIP | SXT | VAN | |
| COPS | 6 | 2 | 0 | 0 | 0 | 3 | 2 | 0 | 6 | 4 | 1 |
| CONS | 4 | 1 | 0 | 0 | 0 | 2 | 2 | 0 | 4 | 2 | 0 |
| Streptococcus spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 10 | 3 | 0 | 0 | 0 | 5 | 4 | 5 | 10 | 6 | 1 |
Abbreviations: FOX, cefoxitin; CFM, cefixime; FEP, cefepime; SAM, ampicillin/sulbactam; IMP, imipenem; GEN, gentamicin; AMK, amikacin; TET, tetracycline; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; VAN, vancomycin; COPS, coagulase-positive Staphylococcus; CONS, coagulase-negative Staphylococcus.
Acinetobacter spp. showed a resistance pattern that the resistance rate to different antibiotics was high. As shown in Table 3 Acinetobacter spp. was resistant to amikacin, ciprofloxacin, cefixime, and gentamicin, with an equal percentage of 70.5%. Klebsiella spp. were highly (75%) resistant to amikacin, ciprofloxacin, cefixime, gentamicin, imipenem, and trimethoprim/sulfamethoxazole but entirely susceptible to tetracycline and ampicillin/sulbactam. Enterobacter spp. mainly (83.3%) resistant to amikacin, gentamicin, and cefixime but completely sensitive to cefepime and showed low resistance to tetracycline and ampicillin/sulbactam (16.6%). Isolated E. coli strains showed the most resistance rate to cefixime (64.7%), followed by amikacin and gentamicin (47%), and no resistance to cefepime and ampicillin/sulbactam.
| Isolated Bacterial Species | The Antibiotics’ Tested Names | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FOX | CFM | FEP | SAM | IMP | GEN | AMK | TET | CIP | SXT | PMB | |
| Acinetobacter spp. | 4 | 24 | 13 | 3 | 20 | 24 | 24 | 3 | 24 | 14 | 0 |
| Escherichia coli | 1 | 11 | 0 | 0 | 4 | 8 | 8 | 1 | 7 | 4 | 1 |
| Enterobacter spp. | 2 | 5 | 0 | 1 | 0 | 2 | 5 | 1 | 3 | 2 | 0 |
| Pseudomonas spp. | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Klebsiella spp. | 1 | 3 | 1 | 0 | 3 | 3 | 3 | 0 | 3 | 3 | 0 |
| Total | 8 | 45 | 14 | 4 | 27 | 42 | 42 | 5 | 37 | 24 | 1 |
Abbreviations: FOX, cefoxitin; CFM, cefixime; FEP, cefepime; SAM, ampicillin/sulbactam; IMP, imipenem; GEN, gentamicin; AMK, amikacin; TET, tetracycline; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; PMB, polymyxin B.
The other issue observed significantly among the isolated bacteria was resistance to multi-drugs. Multidrug resistance (MDR) isolates refer to those bacteria which acquire resistance to at least one agent in three or more antimicrobial groups (13). Among GN-examined isolates -besides Pseudomonas spp. -the rate of MDR was high against different antibiotic categories containing aminoglycosides (amikacin, gentamicin), fluoroquinolones (ciprofloxacin), the third generation of cephalosporin (cefixime), and beta-lactamase (imipenem). Also, among GP isolates, all Staphylococcus spp. were resistant to cefoxitin and identified as methicillin-resistance Staphylococcus, subsequently, all were considered as MDR.
5. Discussion
Abortion is a public health concern worldwide because of its mental and physical side effects, which are occasionally irreparable (14). In Iran, about 80,000 abortions occur annually (15, 16). To follow up on the influential factors in abortion, various criteria have been studied, including demographic characteristics (age, literacy, job, age of marriage, family marriage, etc.) and specific characteristics of abortion, such as the type of abortion, drinking alcohol, smoking cigarette, and drinking coffee; family history of abortion, genetic factors, and anatomic, microbiologic, endocrinologic, immunologic and metabolic factors (17, 18). Due to its significance, studies are recommended to be conducted to determine the probable reasons, focusing on preventing abortion.
Bacteria are one of the etiologic causes of abortion, and more studies are necessary to reveal the association of bacteria with abortion (19, 20). For this purpose, a post-mortem bacteriologic exam can be an optional approach. It seems that post-mortem examination is helpful for two main reasons: (1) determining the etiologic agent of an undiagnosed infection; and (2) confirming an antemortem-diagnosed infection (2, 21). The value of post-mortem bacteriology depends on a thorough autopsy, proper sampling, minimization of post-mortem bacterial translocation, and prevention of sample contamination (22). As mentioned previously, this method is ignored in most healthcare centers or done to a limited degree. Still, this examination is done routinely in a hospital in Shiraz, southwest Iran.
Generally, the frequency of isolated GN bacteria was higher than GP ones regarding the results. Among isolated bacteria, Acinetobacter spp., E. coli, and Staphylococci spp. were the most common isolates in the present study (Figure 1), which was in line with previous studies (6, 23, 24). Amongst GN isolates, Acinetobacter spp., the most isolated bacteria, is the most successful pathogen responsible for nosocomial infections in the modern healthcare system so that it can transmit and colonize quickly in individuals (25, 26). Therefore, studying the history of the mothers to find out if they have experienced hospitalization before an abortion is helpful (27). The ASP results revealed that the most effective antibiotic for GN bacteria was ampicillin/sulbactam (94%), but most isolates (66.6%) were resistant to cefixime. The resistance profile in GN isolates demonstrated notable and worrying since most were MDR; resistance to cefixime (third-generation cephalosporin) has dedicated the highest degree of resistance to itself. After Acinetobacter spp., the Klebsiella spp., Enterobacter spp., and E. coli isolates have exhibited a highly resistant rate against tested antibiotics, respectively; such resistance may be related to their intrinsic resistance mechanisms of having chromosomal and plasmid genes (28).
Among Acinetobacter spp., Acinetobacter baumannii is the most critical species related to nosocomial infections worldwide that readily become resistant to antibiotics by co-existing mechanisms and end up with MDR strains (29), as seen in the results of the current study (27, 30). Twenty out of 34 isolated strains (58.8%) yielded resistance to imipenem, which correlated with significantly increasing carbapenem resistance among A. baumannii globally associated with carbapenem-hydro-lysing enzymes (31). However, it is notable that our study could not detect to which species of Acinetobacter spp. belonged.
Among isolated GP bacteria, Staphylococcus spp. were the predominant isolates justifiable since they are found ubiquitously in nature and frequently on the skin; they are primary causative agents of various types of infection in individuals. Therefore, the mothers can be contaminated easily. Out of 10 detected Staphylococcus spp., six isolates were coagulase positive. Staphylococci spp. isolates had a high degree of resistance to ciprofloxacin, cefoxitin, amikacin, trimethoprim/sulfamethoxazole, gentamicin, and cefixime. According to cefoxitin resistance, the existence of a high frequency of methicillin-resistant Staphylococci spp. (MRSA or MRCoNS) in the present study was confirmed. Given this issue, methicillin-resistant Staphylococci is a severe concern that warrants more attention and precise management. The most effective antibiotic for Staphylococci spp. in our study was tetracycline since all the isolates were susceptible. Moreover, two Streptococcus spp. isolates have been reported as GP, both entirely susceptible to all the tested antibiotics.
The current study was limited by lacking molecular analysis; further investigation using molecular methods for determining the distribution of virulence- and antimicrobial resistance-related genes will propose more information. Also, accessing the history of the mothers of infants can improve the knowledge of the bacteriological role in abortion.
5.1. Conclusions
Most isolates were environmental bacteria with high antibiotic resistance; such bacteria might be considered causative agents of abortion in our region. Therefore, it seems more focus on following the general hygiene of pregnant mothers is essential. However, further evidence of a clinical correlation between aborted fetuses and their mothers is required.