1. Background
A neuropsychiatric disorder known as schizophrenia is characterized by hallucinations, disordered thinking, and cognitive impairment that results in persistent and severe impairment of social and occupational functioning. However, the pathogenesis and cause of schizophrenia are still poorly understood (1). Mammalian physiology, metabolism, and behavior are all controlled by the circadian clock system on a daily basis. This system's molecular mechanisms are made up of several circadian clock genes, the most important of which is circadian locomotor output cycles kaput (CLOCK), which is involved in a wide range of behaviors. Common mental disorders and circadian disorders are linked to CLOCK gene changes (2). Mental disorders frequently accompany disturbances in circadian rhythmic regulation, which typically accompany sleep-wake cycle changes (3, 4). The CLOCK gene has 119238 base pairs long and is located at chr4q12. It regulates other transcription factors' access to DNA and relies on the transcriptional activation of downstream main clock genes to perform its principal function (5, 6). It encodes a protein with 846 amino acids and functions as both a transcription factor and an acetyltransferase (7). Circadian locomotor output cycles kaput protein can form a heterodimer with BMAL1 (encoded by aryl hydrocarbon receptor nuclear translocator (ARNTL)) in order to keep the circadian rhythm. This heterodimer binds to E-box promoter elements upstream of period (PER1, PER2, PER3) and cryptochrome (CRY1, CRY2), which then activate their transcription. In the cytoplasm, heterodimerized complexes of PER and CRY proteins are formed. After the PER/CRY complex reaches the nucleus, it stops the activity of CLOCK and BMAL1, which triggers the transcription of PER and CRY. The PER/CRY inhibitory complex is transferred to the proteasome and destroyed by Casein kinase 1ε phosphorylation and then ubiquitination. Subsequently, a new 24-hour feedback loop is initiated after CLOCK and BMAL1 lose their inhibition (8-10). At the population level, a wide range of inter-individual variations was observed at CLOCK mRNA expression levels in human blood cells (11), demonstrating the diversity of expression regulation across the population. Also, night shift workers' blood cells have the lowest methylation of the CLOCK promoter and the highest expression of CLOCK, indicating a link between CLOCK regulation and circadian rhythm and strengthening the link between epigenetics and circadian rhythm (12-14).
According to the 1,000 Genomes Project Consortium Phase 3 (15), the CLOCK gene's genomic diversity includes 406 SNPs for Africans, 271 for Europeans, 278 for Asians, and 277 for Americans (16). The variant rs6811520T>C in the CLOCK gene is located in the intronic region at position chr4:55449011 (GRCh38.p12) with a minor allele frequency (MAF) T = 0.24 and the variant rs1801260A>G in the CLOCK gene is located in the 3' untranslated region (3'UTR) at position chr4:55435202 (GRCh38.p12) with MAF G = 0.23. The 3'UTR is where the variant rs1801260 is found. This region is crucial to the stability, expression, and function of mRNA. Changes in the intronic region result in cryptic splice sites or exon skipping, which are fundamentally distinct from normal alternative splicing and have the potential to influence the expression of mRNA (17).
According to animal models and in vitro functional experiments, CLOCK is a transcription factor and a histone acetyltransferase, indicating that genetic and epigenetic changes in this gene can contribute to physiological changes that may alter a person's susceptibility to mental disorders. As a result, this study aimed to find out whether the rs1801260 and rs6811520 of the CLOCK gene—a circadian rhythm-regulating gene—underwent genetic changes that may make people more likely to develop schizophrenia.
2. Objectives
This study aimed to investigate the possible relationship between rs1801260 and rs6811520 polymorphisms in the CLOCK gene, as the principal regulator of the circadian rhythm, with susceptibility to schizophrenia in the Iranian population.
3. Methods
3.1. Sampling
The sample population included 103 patients with chronic schizophrenia (46 males and 57 females) and 100 healthy individuals in the control group. Based on the diagnostic and statistical manual of mental disorders (DSM-IV), patients with schizophrenia were selected from Dr. Pasha's Psychiatric Treatment Center and Ganjovian Hospital in Dezful, Khuzestan province. Age and gender were matched between the control and patient groups. The Ethics Committee of Dezful University of Medical Sciences approved this study (IR.DUMS.REC.1399.033). The Declaration of Helsinki serves as the basis for this study. The informed consent form was signed by all patients and the control group.
3.2. DNA Extraction
Blood samples (n = 100 subjects in the control group, n = 103 patients with schizophrenia in the patient group) were collected from unrelated people in the southwest of Iran. The total genomic DNA was extracted from the whole blood using PrimePrepTM Genomic DNA Extraction Kit (from Blood) (Cat No: K-2000, Genetbio Co., Korea), based on the company's protocol. The purity and quantity of genomic DNA were determined using a spectrophotometer.
3.3. SNP Genotyping and Statistical Analysis
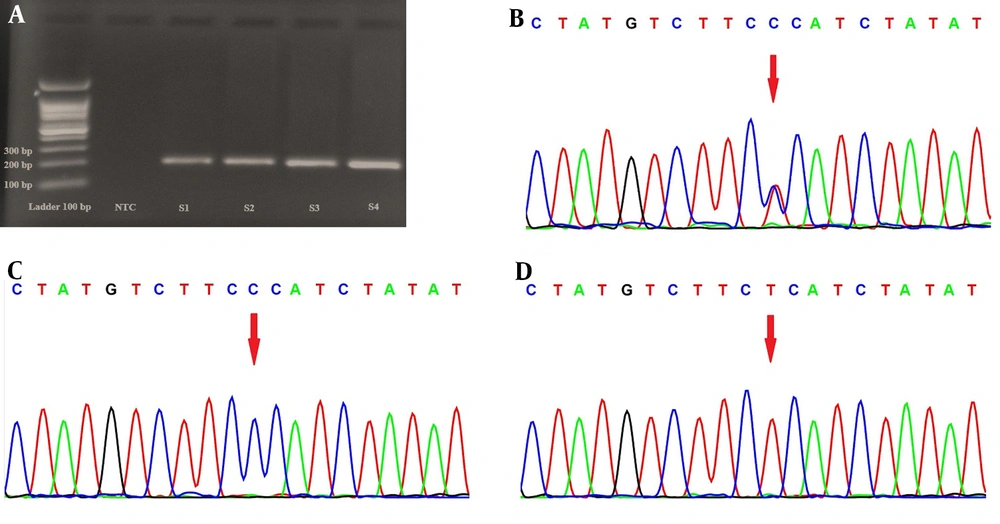
Using dbSNP (short genetic variation) data from the NCBI database, information on CLOCK gene polymorphisms, including rs6811520 and rs1801260, was gathered for this study. Using the Roche LightCycler® 96 (Roche Life Science), the high-resolution melting (HRM) method was utilized to determine the genotype of DNA samples. Twenty µL of the final volume was used for PCR reactions; each reaction consisted of 4 µL of HRM master mix (Solis BioDyne), 2 µL of primer, 2 µL of DNA, and 12 µL of DEPC water. After a five-minute denaturation step at 95°C, the PCR protocol went through 40 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 20 seconds, and HRM at 95°C for 60 seconds, 40°C for 60 seconds, 65°C for 1 second, and 97°C for 1 second, respectively. Different plots were used for the allelic differentiation analysis. The primers for rs6811520 included F: 5'CACTGAAGTGATTCTCATTCCC3' and R: 5'GAGCTTGTTCAAGGTACTGTAGG3' as well as for rs1801260 included F: 5'GAAAATGCTGCCTAGTGCTAC3' and R: 5'TGTCATTTCATAGCTGAGCTTC3'. A product set was created by designing primers with parameters using the Gene Runner and Primer3 software. Additionally, the ABI 3130XL Capillary Sequencing Platform, manufactured by Applied Biosystems/Life Technologies in Carlsbad, California, United States, performed random sequencing assays on 10% of samples to confirm HRM data. The chi-square test (χ2) was used to compare the genotype and allelic frequency between the schizophrenic patients and the healthy control group based on the significance of the difference in odds ratio (OR) and 95% confidence interval (CI). Using the χ2 test, the Hardy-Weinberg equilibrium (HWE) in the groups was also examined. The significance level was set at P < 0.05.
4. Results
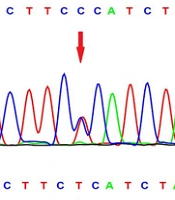
There were 203 participants in the current study who were categorized into two groups consisting of patients with schizophrenia and healthy controls. There were 57 females and 46 males in the schizophrenia patient group, with a mean age of 46.5 ± 1.08 years. Moreover, 56 healthy females and 44 healthy males of the same ethnicity (mean age 38.15 ± 1.3 years) were chosen as the control group. Hardy-Weinberg equilibrium existed among the obtained data. In heterozygous and homozygous conditions, HRM analysis revealed differences in the difference plot at rs6811520 and rs1801260, and Sanger sequencing confirmed HRM results (Figure 1). The frequencies of the genotype and allele of the rs6811520 polymorphism were significantly different in the patients with schizophrenia and healthy control groups (Table 1). The patients with schizophrenia had a higher frequency of the C allele than the control group (83.5% vs. 72%). The CC genotype was associated with an increased risk of schizophrenia (P-value = 0.014, OR = 2.162, 95% confidence interval = 1.161 - 4.028). Additionally, our data indicated that the SNP rs1801260 was not associated with schizophrenia (P-value > 0.05).
| Genotypic Distribution | Allelic Frequency % | P-Value | OR | 95% Confidence Interval | |||
|---|---|---|---|---|---|---|---|
| Genotyps | No. (%) | Alleles | % | ||||
| Schizophrenia | TT | 12 (11.7) | T | 16.5 | 0.014 | 2.162 | 1.161 - 4.028 |
| TC | 10 (9.7) | C | 83.5 | ||||
| CC | 81 (78.6) | ||||||
| Controls | TT | 19 (19) | T | 28 | |||
| TC | 18 (18) | C | 72 | ||||
| CC | 63 (63) | ||||||
Genotypic Distribution and Allelic Frequency of the rs6811520 in the Case and Control Groups
5. Discussion
Psychiatric disorders like major depressive disorder, bipolar disorder, and schizophrenia frequently include descriptions of disorders of the circadian rhythm. The need for circadian assimilation by external factors, which is frequently disrupted in mental illness, and the circadian influence on brain function and mood are among the growing body of evidence suggesting a biological link between mental health and circadian rhythmicity. The phenotypic spectrum of traits related to circadian rhythms and mental health, in addition to the distinct inter-individual differences in this combined susceptibility, suggest that circadian clock genes may also be potential genes for psychiatric disorders (18). The remarkable finding of the present study is that the CC rs6811520 genotype of the CLOCK gene may significantly raise the risk of developing schizophrenia. The rs6811520 variant has been the subject of few studies regarding schizophrenia. Since this variant is in the intronic region, where alterations can cause cryptic splice sites or exon skipping, both of which have an impact on mRNA expression, it may play a role in disease susceptibility. The findings of Abbasi et al.'s study demonstrated a statistically significant difference in the genotype distribution of rs6811520 of the CLOCK gene between multiple sclerosis (MS) patients and healthy control group. Additionally, the TC genotype is associated with an increased risk of MS (19). One of the polymorphisms that has received a lot of attention is the variant rs1801260. It is located in the 3'UTR, which is crucial to the stability, expression, and function of mRNA. Takao et al. conducted a study on 145 patients with schizophrenia and 128 healthy controls to determine whether the CLOCK gene SNP rs1801260 was associated with the condition. There was a significant difference between patients with schizophrenia and healthy controls in both genotypic and allelic frequencies (P = 0.022 and P = 0.015, respectively). Compared to the control group, people with schizophrenia had a significantly higher frequency of the C allele (20). Using mouse embryonic fibroblasts transfected with the rs1801260T/C construct, Ozburn et al. demonstrated that the C allele of the rs1801260 variant resulted in a significant increase in the expression of CLOCK and Per2 mRNA (21). Experiments on a variety of human cell lines have also revealed an increase in CLOCK expression when the C allele is present (22). The functional results may be due to the fact that this SNP is situated in the miRNA-182 interaction site (23). In European and Japanese populations, the rs1801260 variant has been linked to delayed sleep phase syndrome and a preference for activities in the evening (24). The G-protein b3 subunit gene (GNB3) has been linked to the regulation of diurnal preference (25). Sleep duration is also linked to other CLOCK gene polymorphisms (26). In the research conducted by Saleem et al. to determine the specificity of polymorphisms in CAG repeats that encode a portion of the C-terminal region of CLOCK protein, no changes in the length of CAG repeats were observed in Indian-born patients with schizophrenia (27). Clozapine patients treated frequently experience an unpleasant side effect known as sialorrhea, which seems to be related to the circadian rhythm. Four CLOCK gene SNPs were examined by Solismaa et al. in Finnish patients and healthy controls. However, no association was observed in a case-control analysis, and sialorrhea was not identified as a complication in patients with schizophrenia (28). The drawback of the present study may be that it was only observed in Iranian people. As a result, it is suggested that larger, more diverse, and genetically diverse samples should be used for future research.
5.1. Conclusions
According to the findings of the present study, patients with schizophrenia and healthy controls have significantly different frequencies of genotypes and alleles for the rs6811520 polymorphism. We observe that the polymorphism rs1801260 is not associated with schizophrenia. The schizophrenia group has a higher frequency of the C allele of the rs6811520 polymorphism than the control group. The CC genotype may be linked to an increased risk of schizophrenia. As a result, schizophrenia may be associated with CLOCK gene variations.