1. Background
Breast cancer (BC) stands as the most prevalent malignancy affecting women globally (1). BC is a complex and heterogeneous disease with distinct subtypes based on histological features: Hormone-receptor-positive, HER2+, and TNBC (2). HER2+ breast cancer is characterized by amplification or overexpression of the HER2 gene, associated with aggressive tumor behavior and higher mortality rates in the absence of appropriate systemic therapies. Targeted therapies, such as trastuzumab, have shown considerable efficacy in HER2+ breast cancer patients, underscoring the clinical significance of HER2 as a therapeutic target (1).
Long non-coding RNAs (lncRNAs) are crucial regulators in breast cancer. Among these, growth arrest-specific 5 (GAS5) has been extensively studied and described as a tumor suppressor lncRNA with diverse roles in cancer biology. Growth arrest-specific 5levels are decreased in breast cancer tumors, particularly in HER2-positive cases (1). Growth arrest-specific 5exerts its tumor-suppressive effects by promoting apoptosis, inhibiting cell proliferation, and modulating critical signaling pathways in breast cancer, such as the PI3K/AKT/mTOR and PTEN (Phosphatase and tensin homolog) pathways. Dysregulation of these pathways is closely associated with tumorigenesis, metastasis, stem-like characteristics, and drug resistance in cancer cells (1-3).
The PI3K/AKT pathway is central to cancer research, regulating cellular functions including metabolism, growth, proliferation, survival, and protein synthesis. AKT, a key effector of this pathway, influences downstream molecules such as mTOR and other effectors through phosphorylation cascades, thus controlling various cellular processes (4). Approximately 20% to 30% of breast carcinoma cases exhibit HER2 overexpression, resulting in a high relapse rate and reduced overall survival. Trastuzumab resistance has been linked to the presence of breast cancer stem cells (BCSCs) in HER2-positive tumors, with PI3K/AKT and mTOR pathways playing significant roles in regulating BCSC behavior. Dysregulation of these pathways, often involving the tumor suppressor PTEN, a downstream and negative mediator of the PI3K/AKT pathway, has been implicated in trastuzumab resistance (5).
Enhancer sequence alterations have been identified as a common cause of HER2 dysregulation (6). Transcription factors are proteins that bind to short DNA motifs called enhancers. These enhancers play a crucial role in regulating gene expression, and they can exert their effects on target genes even when located hundreds of kilobases away (7, 8). The HER2 gene, located at 17q12, has multiple transcripts, promoters, and enhancers, making it paramount in diagnosing and treating HER2+ tumors (9).
Genome editing tools, particularly the CRISPR/Cas9 system, have revolutionized the field of genetic manipulation, offering precise and targeted modifications in the genome. Leveraging this technology, we employed the CRISPR/Cas9 gene editing method to investigate the functional significance of the putative enhancer region located in the 17q12 (GH17J039694) in HER2+ and HER2- breast cancer cell lines. We assessed the impact of genetic manipulation on HER2 expression, and the expression of other target genes related to apoptosis, proliferation, and stemness. Additionally, we examined the modulation of the PI3K/AKT pathway, a crucial signaling cascade frequently implicated in breast cancer progression. This investigation may better understand the molecular drivers behind HER2 dysregulation and help identify new targets for HER2+ breast cancer treatment.
2. Methods
2.1. Ethical Compliance and Approval
This research adhered to ethical principles and did not involve human participants or the use of human biological samples. Similarly, no live animals were used in any experimentation. The Tarbiat Modares Research Ethics Committee approved the use of commercial cell lines for in vitro experiments as part of a Ph.D. thesis proposal (reference number: IR.MODARES.REC.1399.067, year of approval: 2020).
2.2. Bioinformatics Analysis
The putative enhancer within the HER2 sequence at 17q12 (37,850,592-37,853,472) was analyzed using the UCSC Genome Browser.
2.3. Design of Oligonucleotides
Oligonucleotides for PCR were designed using software and NCBI Primer-BLAST. The primers were synthesized by Metabion (Metabion International AG, Germany). For gRNA design, CRISPOR software (http://crispor.org/ ) was utilized.
2.4. Cloning
The cloning of gRNAs was carried out using the PX459.v2 vector. The cloning process involved the preparation of insert fragments, enzymatic digestion, ligation, transformation, colony check PCR to verify transformed colonies, culture of positive colonies, plasmid extraction, and confirmation of vector inserts through sequencing. Selected vector samples, obtained using a plasmid extraction kit (GeneAll, South Korea), were sent to Rajaei Hospital (Tehran, Iran) for sequencing to confirm the presence of the desired sequence. Detailed information about the cloning procedures can be found in the Zhang Lab Cloning Protocol and the articles by Santos et al. and Ran et al. (10-13).
2.5. Cell Culture
The SKBR3 and MCF7 cell lines were obtained from the Iranian Biological Resource Center (IBRC) in Iran. The cell lines were cultured in DMEM/F12 medium (Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, USA) and 1% penicillin/streptomycin (Bio Basic, Canada). The cells were incubated at 37°C in a CO2-rich environment with 5% humidity.
2.6. Using the CRISPR/Cas9 System, Transfection of Cells and Predicted Enhancer Knockout Can Be Achieved with Cas9 and gRNA Vectors
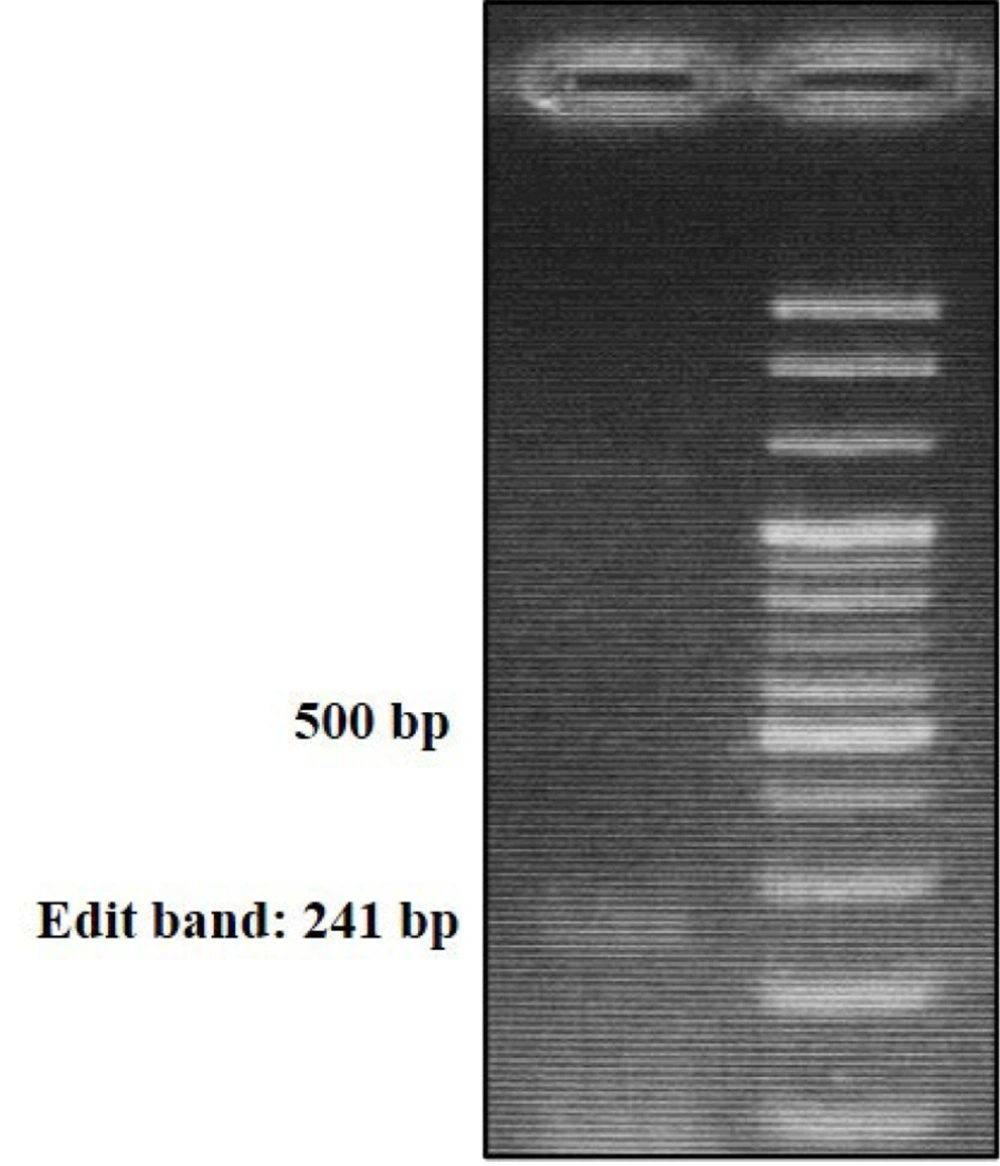
To study the effects of the predicted enhancer on regulation, two gRNAs were designed to target a region of approximately 1000 bp. The goal was to knock out this region using the CRISPR/Cas9 system. The gRNAs were cloned into the vector associated with plate number one. One hundred thousand cells were seeded into each well of a 12-well plate, and gRNA-containing vectors were transfected into cells using Turbofect solution from Invitrogen, USA. After 24 hours, the medium was changed, and the positive control plasmid (PEGFP-C1 (Addgene)) was examined under a fluorescent microscope (Olympus ix53). Selection of cells was performed by treating them with 1.5 μg/mL Puromycin from Sigma-Aldrich, USA, 24 hours after transfection. Pooled cells were subjected to PCR using flanking primers (Edited-Test) to confirm deletion, followed by DNA sequencing at Rajaei Heart Hospital in Tehran. Cellular DNA was extracted using the GeneAll kit from Korea to verify genetic modification.
After PCR-confirmed editing, total RNA from both control and edited cells was extracted using TRIZOL reagent according to the manufacturer's instructions from Thermo Fisher Scientific, USA.
2.7. Gene Expression Assay by Quantitative RT-qPCR
To confirm the inhibition of mRNA expression related to HER2 variants and other studied genes, we performed cDNA synthesis using RNA extracted from transfected and control cells (non-genetically modified parent cells used as control cells to compare the gene expression profiles), according to the manufacturer's instructions (Thermo Fisher Scientific, USA). The synthesized cDNA was used as a template, and GAPDH primers were used as an internal control. The PCR products were analyzed by 1.5% agarose gel electrophoresis. We employed the RT-qPCR technique to measure the transcriptional expression of candidate genes and quantify the cis/trans effects of genomic editing.
2.8. Statistical and Data Analysis
To determine the significance of observed differences in measured variables between edited and non-edited cells, we conducted statistical analyses using the Student t-test and ANOVA. At least two biological replicates were used for all tests, and the reported values are the mean ± standard deviation. We considered P values below 0.05 as statistically significant.
3. Results
3.1. Identification of a Putative Enhancer Region Within HER2
Bioinformatics analysis using the UCSC Genome Browser revealed a putative enhancer region within the HER2 sequence at 17q12. This enhancer region showed potential regulatory significance in the context of HER2 expression.
SKBR3 and MCF7 cells were transfected with Cas9 plasmids and two gRNAs targeting HER2 intron 4 to disrupt enhancer function. Control cells, which were non-genetically modified parent cells, were utilized to compare gene expression profiles through Real-Time PCR.
3.2. Bioinformatics Studies
3.2.1. HER2 and Its Predicted Enhancer Regulatory Region Possess Distinct Features
The HER2 gene consists of 27 exons, producing a 185 kDa transmembrane glycoprotein with 1,255 amino acids (14). The predicted HER2 enhancer is located at position chr17:37850592-37853472 (GRCh37/hg19.2009) and spans 2,881 base pairs. It has been identified as a regulatory region in various databases, including ENCODE (Z-Lab), dbSUPER, and GeneCards (GRCh38/hg38). This enhancer region is found in certain transcript variants of HER2, such as variants 2, 3, and 5, and seems to function as a regulatory region. It starts at intron 3 and extends to the midpoint of intron 4. Notably, this target region predominantly exhibits open chromatin characteristics in embryonic stem cell lines like H1-hESC, H7-hESC, and HePG2 hepatocellular carcinoma (UCSC GRCh38/hg38) (15).
3.2.2. HER2 Enhancer Expression Status
According to GTEX RNA-seq data at UCSC, the mRNA of the predicted enhancer shows significant expression across 54 examined tissues, notably in the nerve, thyroid, skin, esophagus, heart, uterus, and prostate.
3.2.3. Successful Cloning of gRNAs into PX459.v2 Vectors
The cloning process for gRNAs targeting the predicted enhancer region was successfully executed using the PX459.v2 vector. Colony check PCR was performed using the forward primer on the vector trunk (U6F) and the antisense oligo of each gRNA as a specific reverse primer. The positive colonies were then cultured to extract the PX459 plasmid. The accuracy of cloning the gRNAs downstream of the U6 promoter was confirmed by sequencing.
3.2.4. CRISPR/Cas9-Mediated Knockout of Predicted Enhancer Region
Transfection of SKBR3 and MCF7 cell lines with gRNA-containing vectors resulted in the successful knockout of the predicted enhancer region using the CRISPR/Cas9 system. Pooled cells were subjected to Edit-Test PCR (Figure 1) and subsequent DNA sequencing, confirming the targeted deletion.
3.3. Effects of Enhancer Knockout on HER2 Variants and Other Genes
3.3.1. Cis/Trans Effects of Genomic Editing
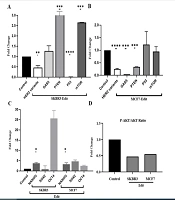
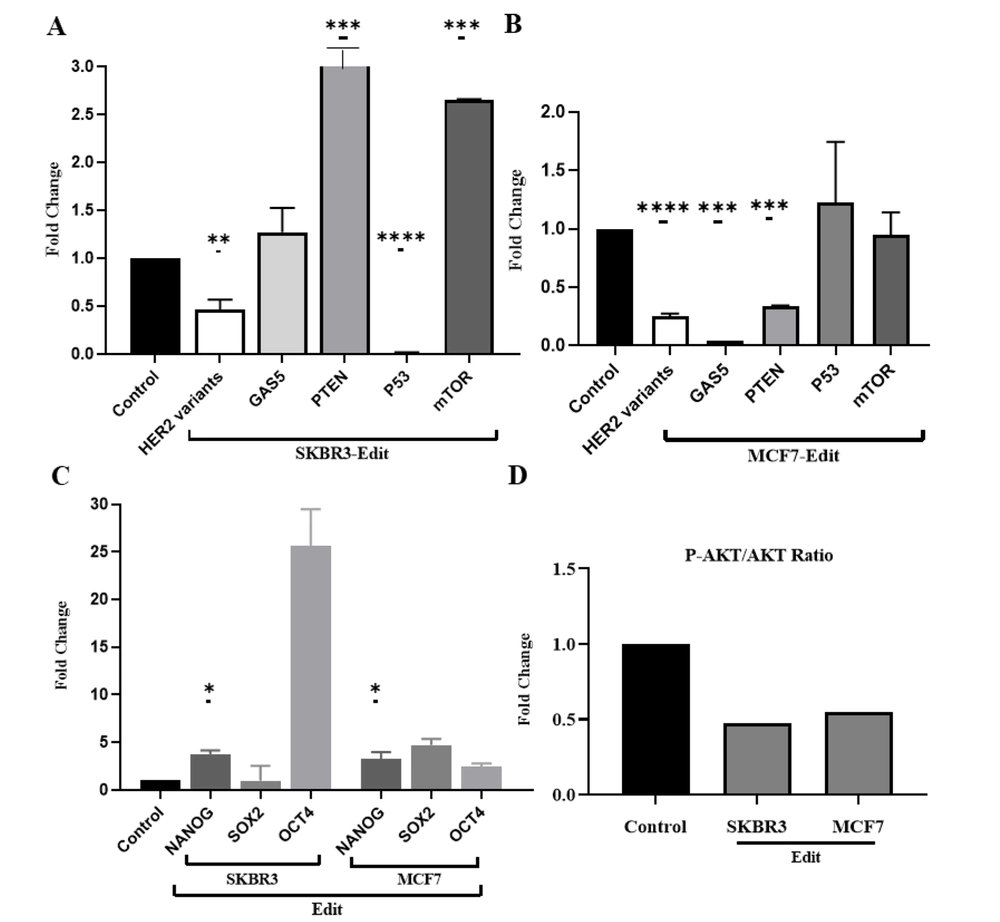
Quantitative RT-qPCR analysis not only confirmed the impact of enhancer knockout on the expression of nearby genes (cis effects) but also revealed potential trans effects on distal genes regulated by the enhancer. Figure 2 illustrates the results of our study on HER2 variants, P53 expression, GAS5, PTEN, mTOR, stemness genes (NANOG, OCT4, and SOX2), and the p-AKT/AKT protein ratio in SKBR3 and MCF7 cell lines after genetic editing.
Illustrates the assessment of the cis and trans-regulatory roles of the studied HER2 enhancer using qRT-PCR. A and B, Panels A and B depict a significant decrease in HER2 variant expression in both cell lines. In SKBR3, the decreased expression of P53 suggests an additional apoptotic pathway, while the notable increase in GAS5 and PTEN expression indicates their role as tumor suppressors promoting apoptosis. Conversely, in the MCF7 cell line, the observations were in contrast, with an increase in P53 aligning with increased apoptosis, while an increase in mTOR and reduced expression of GAS5 and PTEN likely contributes to maintaining the tumorigenic state of the cells. C, Panel C displays the analysis of stemness genes, wherein contrary to expectations, the stemness genes showed increased expression, possibly to sustain reproductive potency. D, Panel D demonstrates that genetic modification significantly reduced the p-AKT/AKT protein ratio (*: P < 0.01, **: 0.001, ***: 0.0001, ****: P < 0.0001). These findings shed light on the intricate regulatory mechanisms involving the HER2 enhancer and its impact on key cellular pathways, offering potential insights for targeted therapeutic strategies.
Firstly, we observed that HER2 variants exhibited reduced expression compared to the control group in both cell lines (Figure 2A and B). Additionally, P53 expression was significantly downregulated in edited SKBR3 cells, while it was upregulated in edited MCF7 cells. Furthermore, our findings revealed a significant downregulation of GAS5 and PTEN in the MCF7 cell line, while in SKBR3 cells, there was an upregulation of GAS5, PTEN, and mTOR (Figure 2A and B).
Additionally, the expression of stemness genes, such as NANOG, significantly increased in both MCF7 and SKBR3 cells. However, the upregulation of SOX2 and OCT4 did not become meaningful (Figure 2C). After genetic editing, the ratio of p-AKT/AKT proteins was reduced in both SKBR3 and MCF7 cell lines, indicating potential alterations in cell signaling pathways (Figure 2D). It should be noted that GAPDH was used as an internal control, and statistical significance was set at P < 0.05.
4. Discussion
HER2+ cancers require accurate diagnosis and targeted treatment, making the identification of markers related to HER2 amplification clinically significant. The main purpose of this study was to use CRISPR/Cas9 technology to explore the role of a potential enhancer, referred to as Her2-Enhancer1, in the HER2 sequence as a regulatory element affecting the transcriptional balance of HER2 variants and other genes, such as the tumor suppressor GAS5 (growth arrest-specific 5) and the AKT/mTOR pathway. Growth arrest-specific 5 ability to inhibit tumor proliferation and induce apoptosis shows promising therapeutic potential (2, 16). Li et al. (as cited in Grossi et al.) identified that GAS5 is downregulated in trastuzumab-resistant HER2-positive BC cells (1). The MYC oncogenic pathway may regulate GAS5 in breast cancer cells, influencing its expression. In cases where GAS5 is silenced, restoring its expression was achieved by inhibiting the PI3K/mTOR signaling pathway. Moreover, upregulating GAS5 increased the G0/G1 arrest ratio and apoptosis in ovarian cancer (2). Growth arrest-specific 5exerts suppressive effects on mTOR, an important PI3K/AKT pathway component, in various cancer types. Additionally, GAS5 positively correlates with PTEN expression, another key PI3K/AKT pathway regulator. Loss of PTEN activity could mediate trastuzumab resistance, suggesting a complex regulatory interplay between HER2, GAS5, and the PI3K/AKT pathway (3).
The knockout of HER2's regulatory region led to a reduction in the expression of its variants in the cell lines studied (as shown in Figure 2A and B). This suggests that this region could be an important enhancer/promoter for HER2. Our previous research (17) supports the finding that gene expressions related to the cell cycle, survival, proliferation, apoptosis, and metastasis were altered after editing. The dysregulation of gene expressions involved in apoptosis, cell cycling, and cell proliferation has been identified as a critical factor leading to uncontrolled cell proliferation and the development of many types of cancers, including breast cancer (18).
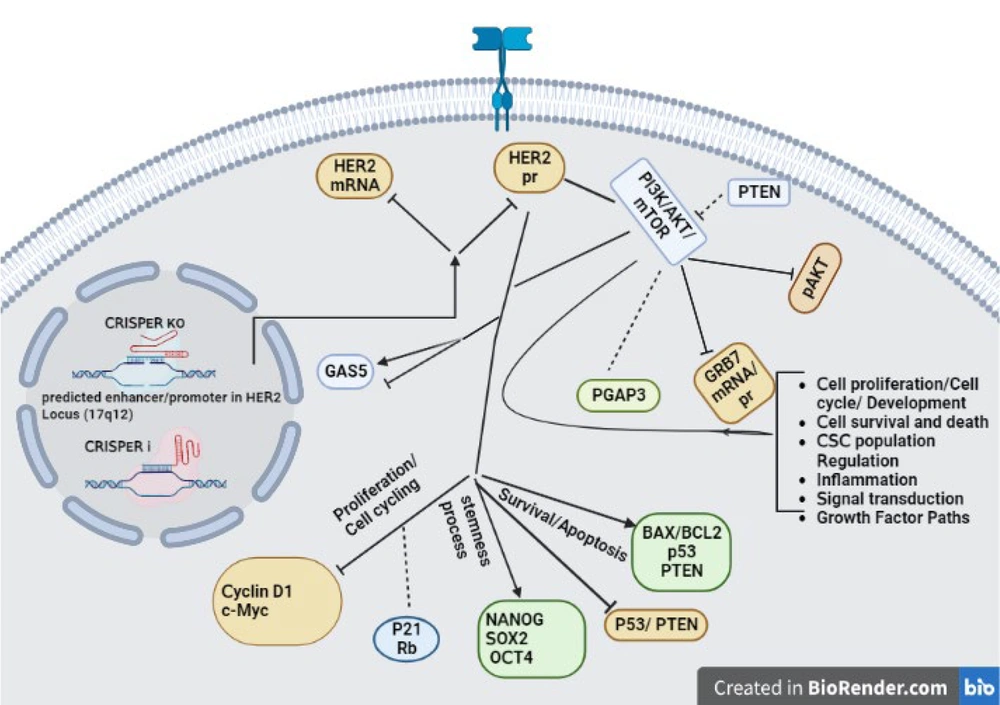
For example, in the edited SKBR3 cell line, the observed reduction of HER2 and its associated protein, as well as the decrease in the p-AKT/AKT ratio, along with the increase in cyclin-dependent kinase inhibitor P21, PTEN, and GAS5 tumor suppressors, and the increase in the BAX/BCL2 ratio, an indicator of apoptosis level, collectively suggest an augmentation of apoptosis through this specific pathway. Furthermore, the decreased levels of the CCND1 oncogene in the edited cells indicate diminished cell cycle progression. Overall, these findings imply a potential regulatory mechanism involving the mentioned factors, which may contribute to an enhanced apoptotic response and impede cell cycle progression in the edited cells.
In contrast, in the edited cells of the MCF7 cell line, the observed decrease in HER2 and its associated protein, as well as the reduction in the p-AKT/AKT ratio, coupled with an increase in the P53 tumor suppressor and a reduction in mTOR levels, causes a noteworthy rise in the BAX/BCL2 ratio. These combined changes suggest the activation of an alternative apoptotic pathway. Additionally, the reduction of CCND1 and c-MYC oncogenes in the edited cells contributes to a decelerated progression of the cell cycle. These findings collectively point towards an alternative pathway of apoptosis induction and an impediment of cell cycle advancement in the edited MCF7 cell line. The alterations in the mentioned factors signify potential regulatory mechanisms that could be driving these cellular responses in the edited cells (Figures 2 and 3).
Schematic representation of Gene Network Associated with HER2 knockout. Gene network analysis reveals genes and signaling pathways affected by a putative HER2-Enhancer knockout (Discussion and Results sections of this article provide more comprehensive explanations and insights into the interrelation of these genes concerning the authors' previous study) (17). Straight arrows: Up-regulation, Inhibitory arrows: Down-regulation, lines: Connections, Dotted lines: Inconspicuous change.
On the other hand, this difference suggests that the edited cells in the two cell lines may undergo apoptosis through different pathways—not the P53 pathway in SKBR3, but potentially through the P53 and mTOR pathways in MCF7 cells. Moreover, the upregulation of GAS5 and PTEN in SKBR3 cells may indicate a potential elevation in the apoptosis rate in these edited cells. In other words, the upregulation of P53 in edited MCF7 cells might affect their apoptotic response. Notably, mTOR showed increased expression in SKBR3 cells, which may be associated with the maintenance of tumorigenicity in these cells (Figure 2A and B).
HER2 expression is notably present in the CSC (cancer stem cell) population, and it plays a significant role in regulating pluripotency via the PI3K/AKT and WNT signaling pathways (19). HER2 and PTEN govern BCSC (breast cancer stem cell) properties. In this study, expression changes of stemness genes NANOG, SOX2, and OCT4 were observed in response to enhancer knockout, possibly indicating the involvement of the enhancer in CSC regulation (Figure 2C). Contrary to our expectation, the results revealed increased expression of all three genes in MCF7 and increased expression of NANOG and OCT4 in SKBR3 cells. Response elements of transcription factors mediating OCT4, NANOG, and SOX2 did not appear in the edited region. Also, other transcripts of their variants likely offset this reduction through other pathways. Further investigations are warranted to unravel the exact molecular events underlying these observations.
In general, although the enhancer knockout impacted HER2 variants, differences were observed in cell survival rates post-editing. HER2 gene expression or copy number variation and compensatory effects by multiple variants could weaken the impact of editing on gene expression. For effective therapeutic strategies to overcome drug resistance and improve clinical outcomes, it's crucial to have a comprehensive understanding of HER2 inhibition and its implications. The results of this study suggest that the region may play a regulatory role as an enhancer in the HER2 gene. Additionally, given its significant role as a tumor suppressor, GAS5 holds promise as a therapeutic target across different cancer types, including breast cancer. Understanding the molecular implications of GAS5 downregulation is essential for developing targeted therapies aimed at reversing drug resistance and improving treatment outcomes.
4.1. Conclusions
The present study provides valuable insights into the regulatory role of the identified enhancer within the HER2 gene and its potential implications in breast cancer biology and therapeutics. The findings highlight the importance of HER2 and its interplay with GAS5 and the PI3K/AKT pathway, shedding light on the complex mechanisms underlying cancer progression and drug resistance. The implications of this research extend beyond breast cancer, with GAS5 emerging as a potential therapeutic target in various types of tumors. Future studies and therapeutic interventions based on these findings have the potential to significantly impact cancer treatment strategies and patient outcomes.