1. Background
Respiratory disorders impose a tremendous burden on global public health. Acute lower respiratory tract infections have been among the leading three reasons for mortality and morbidity among adults and children for decades, indicating the astounding global burden of lung infections (1).
Military trainees are at elevated risk of acute respiratory infections (ARI), disrupting their preparation and contribution to military readiness and causing tremendous losses. Immunity wanes under conditions such as high physical and psychological tension and in crowded places like military training camps (2). Respiratory disorders have been described as the main causes of morbidities and the shortening of training duration among military personnel (3).
Investigations have also shown that key viral etiologies, including adenoviruses, influenza A and B viruses, Epstein-Barr virus, coronaviruses, and rhinoviruses, are mainly associated with ARIs. Human rhinovirus (HRV) is the causative agent of one-half to two-third of common cold cases (4, 7). Infants may be infected 8 to 12 times per year, while adults usually contract the virus 2 to 3 times annually, and multiple peaks may occur during the year (4).
Human rhinoviruses, first characterized in vitro in 1956 (5), belong to the Enterovirus genus of the picornaviridae family and are classified into three species, namely HRV-A, HRV-B, and HRV-C. These viruses impose high medical costs and are responsible for about three out of four cases with upper respiratory tract infections (URTIs) or cold symptoms (nasal obstruction, coughing, rhinorrhea, and sore throat). They are also major causes of the aggravation of chronic obstructive pulmonary infections and asthma. Human rhinoviruses are mainly transmitted by aerosolization. Thus, HRVs inflict remarkable epidemiological, clinical, and economic impacts (e.g., missing a job, an education, or medical treatment) (6). To date, the number of strains/serotypes of HRVs have been reported to be more than 160 (7).
Traditionally, HRVs are characterized by URTIs, otitis media, and sinusitis. Using PCR-based assays, HRV has been identified as an LRTI pathogen, especially in asthmatic patients (children and adults) and immunocompromised hosts. There are currently no licensed antiviral agents for HRV, and therapy is predominantly supportive. Vaccine development efforts have been hampered by the presence of more than 100 HRV serotypes with an extremely high heterogeneity at antigenic sites. The management of HRV infections remains predominantly supportive via alleviating symptoms by over-the-counter remedies (8).
Human rhinoviruses invade the respiratory system’s epithelial cells, especially the nasal passage and nasopharynx, and do not seem to spread to the submucosa, lower respiratory parts, or digestive mucosa since HRVs optimally propagate between 33 and 35°C, and relatively higher temperatures at lower airways restrict HRV proliferation. Certain HRV serotypes, such as HRV-C, uniformly propagate between 33 and 37°C. After RSVs, HRVs are the most common causes of wheezing in babies. Also, HRVs are repeatedly identified in upper airway sputum and bronchial biopsies (9, 10), as well as the lower airway biopsies of children with chronic coughing (11, 12).
2. Objectives
Here, we investigated the prevalence of the serotypes and genotypes of HRVs in military trainees in their mid-20s, who had just been recruited to Iran’s Military Training Camp. Individuals with ARIs were prospectively examined for HRV molecular and epidemiological features throughout the primary military training course.
3. Methods
3.1. Subjects and Sample Collection
From January 2015 to February 2016, many new reservists experienced an intensive three-month essential preparation course, throughout which they remained together in a single-wide barracks within a large dormant quarter. Four hundred respiratory specimens were obtained from the soldiers of the military barracks, who attended the medical center of the preparation camp with the clinical symptoms and signs of an influenza‐like illness (ILI) or URR [such as an oral temperature of ≥ 37.5°C (i.e., febrile respiratory illness (FRI)], sore throat, cough, and runny nose) or fresh ARI (a body temperature of < 37.5°C). Following the medical examination, specimens including sputum, throat swabs, and nasal wash were obtained. Those with a history of severe respiratory disorders such as asthma and chronic obstructive pulmonary disease (COPD), the patients refusing to take antiviral treatments, and military personnel were excluded from the study. Specimens were shipped to the laboratory in virus transport media and stored at -70°C until use.
3.2. RNA Extraction and cDNA Synthesis
Viral RNA was isolated from 200 μL of each sample employing a viral RNA extraction kit (Roche Applied Science, Mannheim, Germany) as directed by the manufacturer. RNase-free-DNase I was used to extract DNA. Using a Nanodrop 2000 spectrophotometer (Thermo Science, the USA), the quantity and purity of nucleic acids were measured. After that, 0.1 ng to 5 μg of the extracted RNA was used to produce cDNA. Then 8.2 μL of the isolated viral RNA was explicitly used in cDNA synthesis using a first-strand cDNA synthesis package (Thermo Scientific RevertAid) and random hexamers as instructed by the manufacturer. Consequently, 2 μL of the synthesized cDNA was applied for conducting PCR.
The online Primer 3 platform was employed to design primers. Two oligonucleotides [P-Forward (5'-CAAGCACTTCTGTTTCCC) and P-reverse (5'-CACGGACACCCAAAGTAGT)] were applied to amplify a 397-bp fragment in the 5-UTR region of HRV genome. The specificity of the primers used in this study was confirmed before conducting PCR.
The PCR mixture (25 μL) contained 2.5 U pfu polymerase (Fermentas Life Sciences, Vilnius, Lithuania), 2.5 μL of 0.2 μM primers, 10 X buffer, and 200 μM dNTP. Afterwards, PCR started with an initial denaturation phase at 95°C for 2 min, followed by 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min. For the final extension, reaction tubes remained at 72°C for 5 min. Amplicons were run on 2% agarose gel prepared by TAE buffer and visualized by ethidium bromide staining. A 100-bp DNA ladder [marker XIV (Roche Applied Research, Mannheim, Germany)] was utilized as a molecular weight marker.
3.3. Nucleotide Sequencing and Phylogenetic Analysis
The 5-UTR amplicons of HRV genome were sequenced, and the findings were compared to the Query Sequence via Clustal X Software (version 1.83) and evaluated using the BioEdit and Treecon programs for drawing a phylogenetic tree. This software has a Kimura Nj system with a bootstrap value of 1000 Rep. A phylogenetic tree with a bootstrap of > 70% is considered to be accurate.
4. Results
A total of 400 military trainees showing the clinical symptoms of RIs were enrolled in this research. All the subjects were male with an average age of 21 ± 4 years. The most common blood groups were A (141/400, 32.2%) and O (139/400, 34.7%), and the least frequent was the AB blood group (34/400, 8%). Regarding clinical symptoms, most patients complained of sore throat (n = 233, 58.25%) followed by cold (n = 41, 10.25%), cough (n = 33, 8.25%), and rhinorrhea (n = 10, 2.5%) (Table 1). Regarding residence in the camp, 51% (205/400) went outside every day, and 49% (195/400) remained in the camp for a long period.
| Symptoms | HRV PCR | Total | |
|---|---|---|---|
| Negative (%) | Positive (%) | ||
| Fever > 38.5°C | 1 (0.2) | 0 | 1 (0.2) |
| Cough | 31 (7.7) | 2 (6.8) | 33 (8.2) |
| Rhinorrhea | 9 (2.2) | 1 (3.4) | 10 (2.5) |
| Shortness of breath | 4 (1) | 0 | 4 (1) |
| Sore throat | 217 (54.2) | 16 (55.1) | 233 (58.2) |
| Sore throat/cough | 6 (1.5) | 0 | 6 (1.5) |
| Sputum/cough | 3 (0.7) | 0 | 3 (0.7) |
| Sore throat/cold | 36 (9) | 2 (6.8) | 38 (9.5) |
| Sore throat/headache | 3 (0.7) | 1 (3.4) | 4 (1) |
| Sore throat/rhinorrhea | 3 (0.7) | 0 | 3 (0.7) |
| Sore throat/cough | 20 (5) | 2 (6.8) | 22 (5.5) |
| Cold | 36 (9) | 5 (17.2) | 41 (10.2) |
| Sore throat/fever | 2 (0.5) | 0 | 2 (0.5) |
| Total | 371 (93) | 29 (7) | 400 (100) |
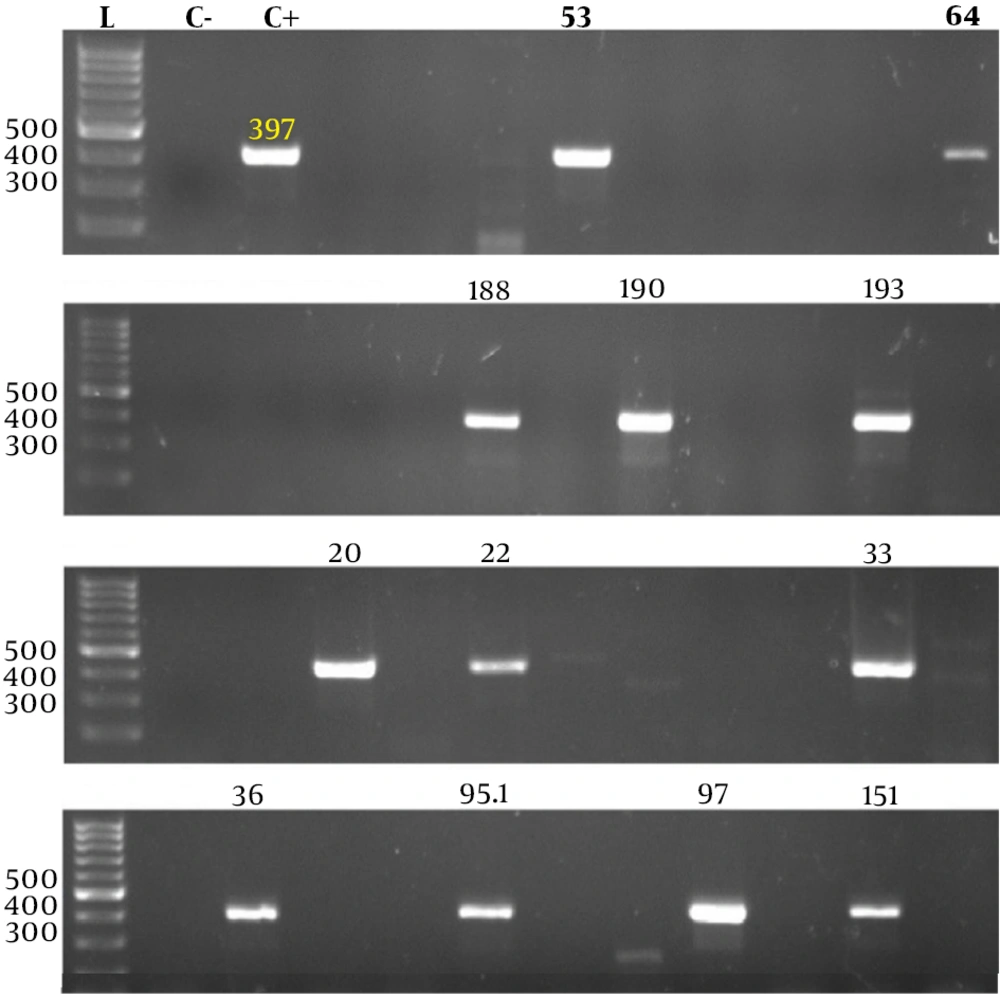
The amplification of the unique HRV 5'-UTR region produced a sharp 397-bp band (Figure 1). Totally, 29 patients (7%) aged 22 ± 5 years (95% CI: 20 - 24) had HRV infection (P = 0.0001) based on RT-PCR findings. Of these, 19 (59%) had a sore throat, five had flu, three had sore throat and cold, and two had a cough (P = 0.0001). Regarding ABO blood groups, 46% (15/29) had O blood group followed by A (7/29, 21%), AB (5/29, 15%), and B (2/29, 6%) (P = 0.0001). Of these, 71% (23/29) were travelers and settled out of the camp at night, and 18% (6/29) remained in the camp for a long period (P = 0,081).
HRV RT-PCR. Products were visualized by agarose gel electrophoresis. Lanes 53, 64, 188, 190, and 193 show 397-bp bands and belong to HRV positive cases. A 100-bp DNA marker (Roche Applied Science, Mannheim, Germany) was used to designate the band’s size (lane L). C-, negative control; and C+, positive control.
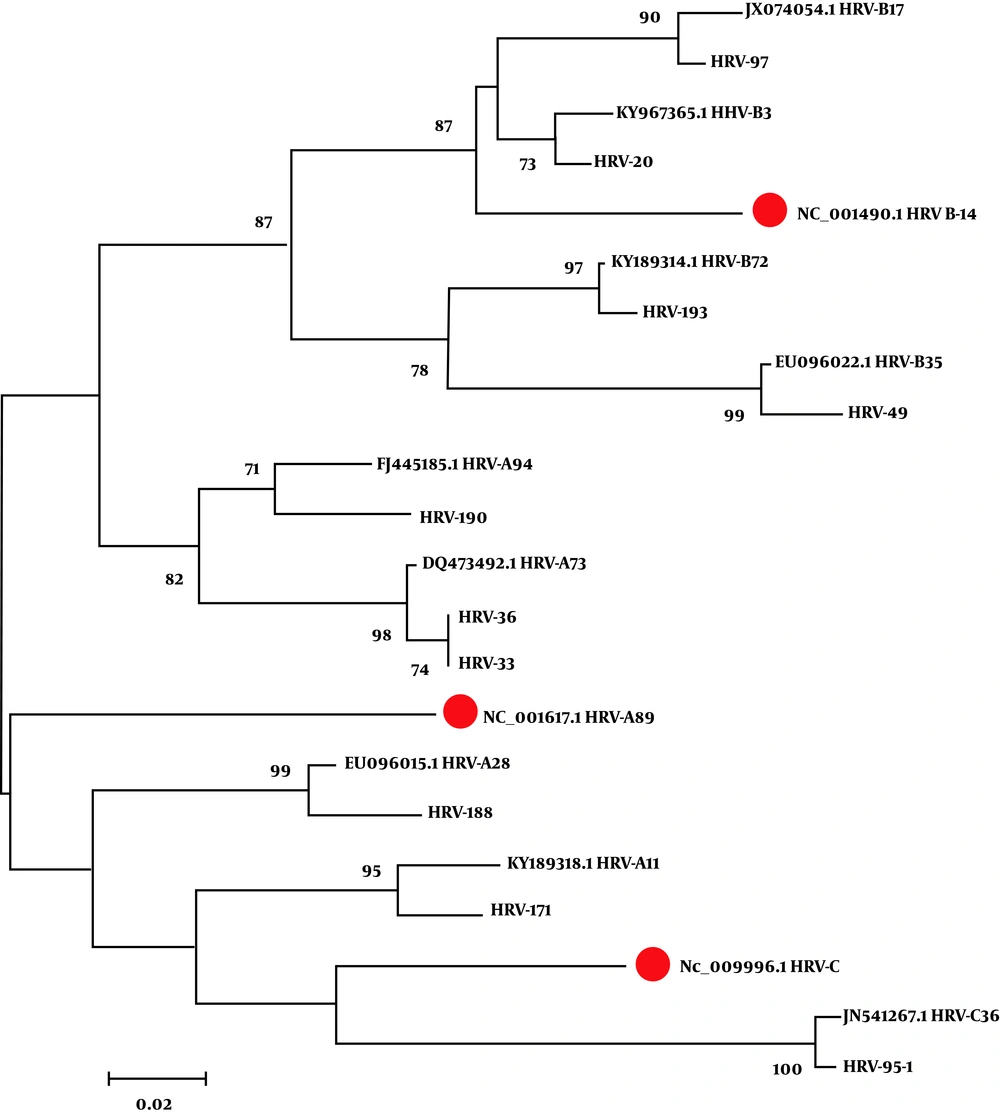
Ten out of the 29 positive samples were sent for nucleotide sequencing and phylogenetic analysis. The existence of HRV was confirmed in the samples tested based on nucleotide sequencing. The phylogenetic tree revealed 5 (50%) HRV-A, 4 (40%) HRV-B, and 1 (10%) HRV-C. Of patients with HRV-A infection, 80% had a sore throat, 10% had a sore throat with the cough, and the remaining had cold symptoms. In HRV-B positive patients, only a sore throat was found. Regarding residency inside or outside the military base, 51.2% went outside the camp, and 48.8% had a permanent presence in the base. Of those with HRV-A, most (n = 7; 3.6%) were inside the camp, and the remaining (n = 3; 1.5%) were outside the camp. There was also a positive HRV-B specimen among those who migrated outside the camp. Of patients with HRV-A infections, three (30%) had < 19 years, 4 (40%) had 20 to 21 years old (n = 4), one (10%) belonged to a 43 years old man, and one (10%) was over 51 years old. Patients with HRV-B infections were 20 to 21 years old.
The phylogenetic examination of the sequenced RT-PCR products exhibited that strains No. 36 and 19 were compatible with the strains isolated from the Philippines and the USA, respectively. The majority of the isolated strains were comparable to the sequences obtained from the USA (Figure 2).
The phylogenetic tree drawn using the neighbor-joining method for some HRV-positive samples. The strains identified in this study and out-group sequences were analyzed against reference sequences (red circles). Phylogenetic trees were constructed using the neighbor-joining method (Kimura’s two-parameter) with 1,000 bootstrap values. The numbers in each branch indicate how close strains were to each other.
5. Discussion
Screening HRVs, particularly in severe cases of RTIs, may be accompanied with the detection of new emerging viral strains, endangering global health security. The precise detection of pathogens in the clinical setting is critical for early interventions and avoiding disease propagation (13, 14). Pneumonia is the major cause of morbidity and mortality in children worldwide, with nearly 120 million cases diagnosed each year (15). Respiratory viruses are responsible for 14 to 80% of childhood community-acquired pneumonia in infants, with HRV being the most commonly identified pathogen. Various HRV strains (i.e., A, B, and C) have been reported in association with mild RTIs and asthma exacerbation (16).
Militaries are particularly and theoretically at higher risks of RTIs than other social classes. In a survey conducted by the Acute Respiratory Diseases Commission in the US recruitment camps, the prevalence of RTIs among recruited trainees was greater than that of other trained military groups (17). Experiments in military applicants also reported elevated rates of RTIs. Among military trainees, HRV infections were related to RTIs with more difficulties in breathing and a lower rate of pneumonia (18). In young and healthy individuals, these pathogens typically cause moderate and self-limiting infections (19). Studying the clinical and/or epidemiological features of HRV-infected trainees indicates the emergence of the "common cold" syndrome (50%) and other signs such as headache and sore throat and a surge in the incidence of this infection among trainees (20 - 70%) with LRT signs such as dyspnea and pneumonia (20).
Tan et al., in their investigation, reported that the prevalence of HRV infection was between 3 and 4% in USAFSAM Military Treatment facilities (MTFs) (18). The routine monitoring of febrile respiratory infection (FRI) cases by Singaporean military personnel from May 2009 to October 2012 revealed an incidence of 7.4% for rhinoviruses (18). Radin et al., in their study on the epidemiology of RTIs among three US communities (citizens along the US-Mexico frontier, recipients of the Department of Defense (DoD), and military recruits) between October 2011 and March 2013, identified rhinoviruses in 16% of samples. In addition, the disease was more severe in outpatient FRI cases than in inpatients (SARI) (21). Another study conducted by Lau et al. on the incidence of respiratory viral infections in Singapore military members in 2016 exhibited that rhinoviruses were the most prevalent pathogens detected in respiratory specimens (nearly 47% of samples) (3). Levy et al. investigated the incidence of upper RTIs among novice trainees in military camps in Thailand and identified rhinoviruses in 22 (28.57%) out of 77 nasal/throat swab specimens obtained during the basic training period (17). Wang et al. assessed throat swabs from military recruits with FRIs by microarray and showed that out of 97 specimens, 78 were HRVs (73 cases of HRV-A and five cases of HRV-B). Also, 46 of 73 confirmed HRV-A specimens were positive for HAdV-4 (22). Another study by Yun et al. on acute RTIs among the military trainees vaccinated against adenoviruses in Philadelphia from June 2008 to August 2013 revealed a dramatic reduction in the incidence of the infection compared with before vaccine usage, all viral infections identified in 68.4% (1266/1850) of which 17.8% (335/1880) were rhinovirus infection (23).
Furthermore, Kopra et al. searched for HRV species in the sputum specimens of military trainees with respiratory infections and exhibited that among 386 sputum specimens, 146 (37.8%) were positive for HRVs based on RT-PCR, including Of 146 HRV positive strains, 55 were positive in typing assay, and then, 29 had a reliable sequence for interpretation among 29 strains, 18 were HRV-A, 5 were HRV-B, and 6 were different genetic types (16).
In this research, 400 patients were recruited, of whom 29 were diagnosed with HRVs. Of the ten samples sequenced, five, four, and one rendered HRV-A, HRV-B, and HRV-C, respectively, showing similarities in the phylogenetic analysis. The sequences submitted from the Philippines and the US strains in the GeneBank database indicate a high heterogeneity in HRV-A strains. These viruses were not linked with any neurological symptom or mortality in the studied population. There were no significant differences in the clinical and demographic features of the patients infected with various HRV strains. Many studies have indicated that HRV-A is a minimally pathogenic virus. Disease severity was not significantly different between patients with HRV-A and HRV-C. The prevalence of the O blood group was high among trainees with HRV infection, suggesting a role for this blood group in susceptibility to this virus.
From the limitations of this study are relatively low sample size (especially the positive cases) and not discriminating ARI from FRI cases. In fact, any ARI case may eventually develop FRI. This limitation might have contributed to the underestimation of FRI events. This study primarily included young adult males in a semi-closed military environment. Thus, the results recorded here would not be relevant to the general adult population (24).
5.1. Conclusion
This investigation aimed to describe the prevalence, clinical features, and molecular epidemiology of HRVs in Iranian military trainees with RTIs. Our findings showed that most RTIs were due to HRVs, particularly HRV-A. Nevertheless, most of our samples had been collected during the first six months of the year, so it is recommended to perform comparative and more comprehensive research in the second half of the year, during which these diseases are more prevalent. Studies in the military barracks of other regions of Iran will help to determine the actual prevalence of HRVs and their possible sources in the country. Despite advances in molecular diagnostic methods, allowing the rapid detection of HRV infections, antiviral agents, and vaccination remain significant prerequisites for the management of these infections.