1. Context
Polymerase chain reaction (PCR) involves the use of molecular procedures to amplify minute quantities and produce several copies of genetic materials (DNA and cDNA). It is a powerful method used for DNA amplification, and its utilization has been approved in detecting minute amounts of viral RNA in cells and tissues (1). At present, RT-PCR is one of the most sensitive procedures for detecting mRNA and quantifying SARS-CoV-2. It can precisely evaluate small quantities of genetic materials via producing cDNA from RNA by reverse transcription and subsequently amplifying cDNA by PCR (2). Among the diagnostic methods of COVID-19, RT-PCR (most reliable) and rapid Ag-Ab tests (with still questionable specificity and sensitivity) are most widely employed. Real-time reverse transcriptase–polymerase chain reaction using nasopharyngeal swabs has been proven to be highly effective for detecting the virus in clinical samples and confirming a diagnosis (3).
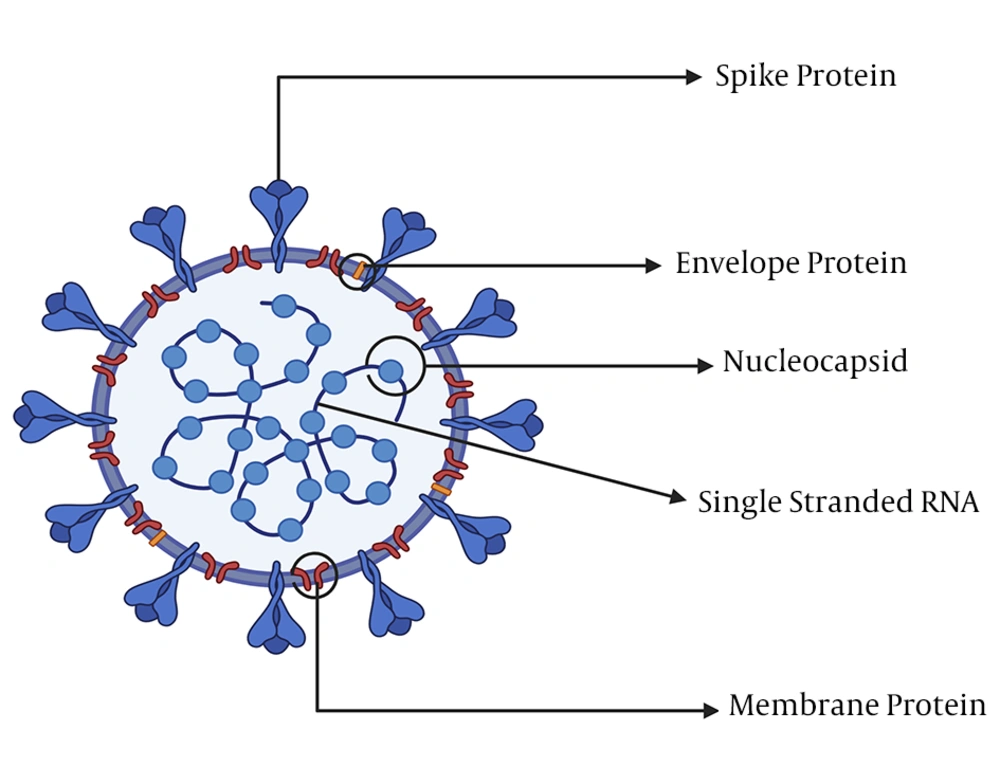
Towards the end of December 2019, a new coronavirus strain emerged in Wuhan, a city in China, with the ability to be transmitted from humans to humans. The disease is characterized by severe pneumonia symptoms resulting in the death of many people. Coronaviruses belong to the phylum incertae, order Nidovirales, family Coronaviridae, genus Betacoronavirus, and subgenus Sarbecovirus (4). These viruses are positive-sense and single-stranded RNA organisms (5) and have spikes, a membrane, an envelope, and nucleocapsid proteins (6). Coronaviruses can fall into one of the four subgenera: α-CoVs, β-CoVs, γ-CoVs, and Δ-CoVs, based on their genetic and antigenic materials, and their genome sizes vary from 26 to 32 Kbp, the largest genome for RNA viruses so far (7).
The SARS-CoV-2 virus was officially reported to the world health organization (WHO) office in China on 31 December 2019 as a pneumonia of unknown etiology (8). Previously known as the novel coronaviruses (2019 nCoV), it was later renamed the coronavirus disease 2019 (COVID-19). In severe cases, the virus causes complicated respiratory illnesses (5, 9). The SARS-CoV-2 virus differs from Middle East respiratory syndrome coronavirus (MERS-CoV) and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) as it often exposes patients to severe respiratory pneumonia (5, 10). The disease has spread globally, mostly through human-to-human transmission.
Military organizations like civilian communities are also under considerable threat due to COVID-19 and other emerging infectious diseases transmitted through human-to-human interaction. Amid its global spread, COVID-19 may affect military training and operations, thereby necessitating military preparedness to counteract any form of the pandemic threatening military personnel. Armed forces are unique due to their functions in territorial defense of nations, and therefore, bioterrorism and bio-warfare often use pathogenic microorganisms to target the military and weaken its structure. For this reason, there is often a need to carry out disease surveillance in armed forces using molecular techniques that will guarantee the safety of all and prevent any form of outbreaks. The U.S. army has been at the forefront of the fight against the COVID-19 pandemic and has played a prominent part in mobilizing medical assistance and constructing emergency facilities to cope with the predicted outbreak (11). Germany has mobilized 15,000 troops to help city officials address the pandemic, while Poland, for example, has enabled thousands of soldiers to patrol locked streets, disinfect hospitals, and reinforce border control (12). Military operations have been curtailed or, in some cases, suspended in France, Italy, and Spain, the nations that were hardest hit by the outbreak (12). The aim of this review was to discuss the applications of RT-PCR for detecting and screening SARS-CoV-2 among armed forces and the successful halting of COVID-19 transmission.
2. Origin of SARS-CoV-2 and Symptoms of COVID-19
Several reasons demonstrated that SARS-CoV-2 was of zoonotic origin (10, 13). A report suggested that early-infected patients were exposed to the local animal market or seafood market in Wuhan, China (5), suggesting possible animal-to-human transmission. Earlier reports from the Middle East indicate the transmission of MERS-CoV from the camel to human (13, 14). With the global spread of the virus, it is now clear that human-to-human transmission (15, 16) remains the principal transmission route of the virus. Viral genome sequencing shows a high similarity between SARS-CoV-2 and the bat coronavirus (9, 17, 18). Upon the entry of the virus, the infected person may be normal, presenting no symptoms. The symptoms ranging from mild to severe, including fever, fatigue, shortness of breath, sore throat, runny nose, sneezing, and pneumonia, may manifest two to 14 days after exposure to the virus (17, 18).
3. Structural and Genomic Organization of SARS-CoV-2
Coronaviruses are large viruses with an approximately 30 kb plus-stranded RNA and about 125 nm diameter (19) that are usually capable of infecting a wide range of animals, including human beings (20). The SARS-CoV-2 virus consists of an enveloped, single, positive-stranded genomic RNA encoding nearly four (4) viral structural proteins, viz. spike (S), envelope (E), membrane (M), and nucleocapsid (N) (Figure 1) 3 - 5 proteins, following the characteristic gene order [5′-replicase (rep), spike (S), envelope (E), membrane (M), and nucleocapsid (N)-3′] (21). The nonstructural proteins are encoded by the rep gene making up about two-third (2/3) of the genome at the 5′ end. The S glycoprotein facilitates receptor binding and the subsequent entry of the virus into hosts’ target cells and is thus a significant therapeutic target (22, 23). The proteins M and E play important roles in the assembly of viral particles, and the protein N is required for the synthesis of RNA (19).
The virion’s most abundant structural protein is the M protein that is about ~25 - 30 kDa, consisting of three transmembrane domains (24) and maintaining the virion’s shape. The N-terminal is a small glycosylated ectodomain, while the C-terminal is a much larger endodomain extending 6 - 8 nm into the viral particle (25). Though M proteins are co-translationally integrated into the ER membrane, almost all lack a signaling sequence. Reports indicate that the M protein exists in the virion as a dimer and may exhibit the configurations permitting it to foster the membrane curvature and bind to the nucleocapsid (26). Within the virion, the E protein (proximately 8 - 12 kDa) exists in minute quantities and shows a highly divergent but comparable architecture (27). Both the N-terminal ectodomain and the C-terminal endodomain of the E protein are located in an ion channel. The recombinant viruses lacking the E protein remain not always lethal (28). The E-protein fosters the virus’s assembly, release, fitness, and pathogenesis (29).
The N protein consisting of two domains, the C-terminal and N-terminal domains, is the only protein contained within the nucleocapsid. The protein’s C- and N-terminal domains are able to bind to RNAs in vitro (19). Both the C- and N-terminal domains contribute to optimal RNA binding (30, 31). The frequent phosphorylation of the N protein (32) initiates a structural change that intensifies its affinity for viral versus non-viral RNAs (19). Specific RNA substrates for the N protein include the transcriptional regulatory sequences and packaging signals (33). The N protein binds RNA via a bead-on-a-string type interaction, whereas the packaging signal in the genome (34) binds the RNA binding domain (19). The N protein also binds to a key component of the replicase complex, the nonstructural protein-3 (31, 35), in addition to the M protein (36). Perhaps these protein-protein interactions will help the viral genome to bind to the replicase transcriptase complex and then bundle the encapsidated genome into viral particles (19).
4. Applications of RT-PCR for the Diagnosis of Acute Respiratory Infections
Real-time RT-PCR is commonly used for identifying causative viruses in the respiratory secretions of patients with acute respiratory infections (Figure 2) (37). Viral genetic materials are initially isolated from respiratory specimens and then subjected to RT-PCR via the synthesis of cDNA. Earlier reports have confirmed the use of RT-PCR-based technologies in public health laboratories for managing international outbreaks (38-41). The emergence of COVID-19 has highlighted the dominant application of real-time RT-PCR for diagnostic purposes, and the method remains the principal tool for detecting the virus’s various strains among many existing diagnostic methods (42). It has become instrumental not only in the diagnosis but also in the treatment of COVID-19 (43). Although RT-PCR has a high sensitivity and reliability, the procedure may disrupt the viral genetic material, causing false-negative results in some cases (43). The outbreaks of acute respiratory illnesses are often considered health priorities during military postings, deployments, and operations (44). Influenza and acute respiratory infections can significantly impair military operations (44) and can be used as a pathogen(s) or bio-warfare to suppress enemies during war confrontations. The value of RT-PCR lies in its ability to detect RNA viruses in a given specimen, amplify target genes, and measure viral loads or monitor responses to antiviral drugs (45). By merging both quantification and detection of RNAs from pathogens within the shortest possible time, RT-PCR protocols become a dominant technique in molecular medicine and diagnostic laboratories (46).
5. COVID-19 Surveillance in the Military
Infectious disease surveillance in the military aims to monitor, reduce, control, and prevent outbreaks. Specifically, COVID-19 surveillance may involve the finding of active cases, isolating infected patients, contact tracing, and quarantine. The symptoms of the COVID-19 diseases, including fever, fatigue, shortness of breath, sore throat, runny nose, sneezing, and pneumonia-like symptoms, should be monitored among military personnel. This has been proven to be very effective for identifying COVID-19 cases in many countries. Observing good personal hygiene (washing hands and covering nose when sneezing and coughing), sanitizing public utilities such as door handles, stair rails, as well as providing health education to armed forces may help to contain such diseases in military organizations (47). Laboratory-based surveillance often plays a key role in disease prevention via detecting sources of pathogens, contributing to a more reliable risk assessment of the infectious disease (44, 48). Such knowledge can also be useful in formulating successful prevention methods and providing care to deployed military personnel. Military laboratories should be able to detect pathogens early enough to prevent unnecessary outbreaks and issue timely warnings. Multi-drug resistant pathogens should also be detected in military hospitals and communities.
6. The Specimens Used for COVID-19 Detection
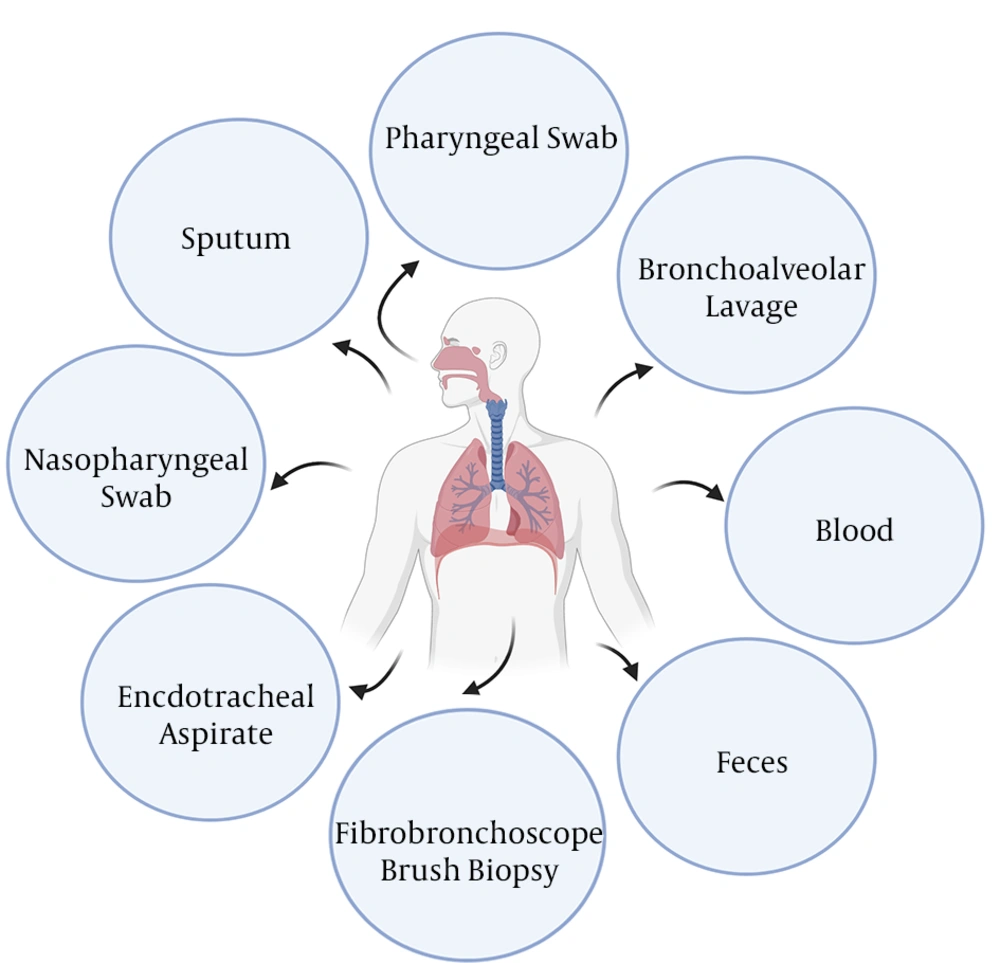
Sample(s) can be obtained from the suspected patient(s) or those who have had close contact with confirmed patient(s). The specimens to be collected, according to the WHO, include upper respiratory specimens (pharyngeal/nasopharyngeal swabs) and/or lower respiratory specimens (bronchoalveolar lavage sputum/endotracheal aspirate) (49) (Figure 2). An investigation on the types of clinical specimens from 1070 COVID-19 patients for the detection of SARS-CoV-2 revealed that bronchoalveolar lavage fluid specimens rendered the highest positive rates (14 out of 15 patients, 93%), followed by sputum (72/104, 72%), nasal swabs (5/8, 63%), fibrobronchoscope brush biopsy (6/13, 46%), pharyngeal swabs (126/398, 32%), feces (44/153, 29%), and blood (3/307, 1%). None of 72 urine specimens tested positive (50). Figure 2 shows the possible specimens that can be used for detecting COVID-19.
7. Principle of RT-PCR Procedure
Polymerase chain reaction consists of three stages; denaturation, annealing, and extension. In the denaturation phase, the reaction temperature is raised up to 95°C, which melts double-stranded DNA into a single-stranded molecule via disrupting hydrogen bonds between complementary bases. In the annealing phase, the temperature is decreased to around 45 - 60°C that is far below the melting temperature (Tm) of primers to allow them to bind to the template. Finally, in the extension process, the temperature is raised to 72°C, which is optimal for DNA polymerase activity to allow hybridized primers to expand.
The polymerase chain reaction is an advanced technique providing a fast and accurate method to amplify DNA and allowing gene cloning and manipulation in biomedical research. This procedure has facilitated the diagnosis of infectious diseases and cancers based on genetic features. The use of reverse transcriptase to determine RNA levels and real-time DNA amplification and quantification are among advanced applications of PCR (51). In this technique, specific probes/primers, which are used to detect target viral genes, are key elements. For SARS-CoV-2 detection, primers and probes should be chosen from the virus’s E, RdRP, and N genes. For example, the primers and probes distributed by TIB MOLBIOL are used in most countries to detect the presence of SARS-CoV-2 via amplifying E, RdRP, and N genes. These can be used as controls for RNA extraction or PCR. An example of an internal control RNA includes the Equine Arthritis Virus genomic fragment, β actin. The genetic material of the virus (RNA) is initially extracted from the sample and reverse transcribed to cDNA, which is then amplified using a real-time PCR machine. As the reaction progresses, the probe anneals to a viral target sequence for which the two primers (forward and reverse) are specific. In the RT-PCR phase, Taq polymerase, a highly thermo-stable enzyme, eliminates the probe by its nuclease activity, separating the quencher dye from the reporter dye, thereby producing a fluorescent signal (52), which is then detected at each cycle.
8. Specific Primers for Detecting SARS-CoV-2
Primers are short-stranded pieces of nucleic acid, annealing to a region of the genetic material to be amplified. One of the most important factors influencing the success and quality of quantitative real-time PCR is the design of primers and probes. In fact, precise and consistent quantification depends on the use of effective primers and probes (53). For longer PCR products, selecting efficient primers becomes more difficult. To design effective primers, it should be kept in mind to avoid the formation of primer-dimers, minimize self-complementarity, and not using primers with too low (melting temperature) Tm and/or internal stability (54). Several different primers have been used to detect SARS-CoV-2 in different respiratory samples (37, 55-60). These primers are very specific for viral genes, annealing to target genes and allowing SARS-CoV-2 detection. The common primers used for detecting SARS-CoV-2 are presented in Table 1.
| Target Genes | Intended Purposes | Country | Forward Primer | Reverse Primer | Probe | Reference |
|---|---|---|---|---|---|---|
| N gene | COVID-19 | Thailand | CGT TTG GTG GAC CCT CAG AT | CCC CAC TGC GTT CTC CAT T | CAA CTG GCA GTA ACC A | (36) |
| N gene | COVID19 | Japan | AAA TTT TGG GGA CCA GGA AC | TGG CAG CTG TGT AGG TCA AC | ATG TCG CGC ATT GGC ATG GA | (37) |
| N gene | COVID-19 | China | GGG GAA CTT CTC CTG CTA GAA T | CAG ACA TTT TGC TCT CAA GCT G | TTG CTG CTG CTT GAC AGA TT | (38) |
| N1 gene | COVID-19 | USA | GAC CCC AAA ATC AGC GAA AT | TCT GGT TAC TGC CAG TTG AAT CTG | ACC CCG CAT TAC GTT TGG TGG ACC | (39) |
| N2 gene | COVID-19 | USA | TTACAA ACA TTG GCC GCA AA | GCG CGA CAT TCC GAA GAA | ACA ATT TGC CCC CAG CGC TTC AG | |
| N3 gene | COVID-19 | USA | GGG AGC CTT GAA TAC ACC AAA A | TGT AGC ACG ATT GCA GCA TTG | AYC ACA TTG GCA CCC GCA ATC CTG | |
| RdRp /Orf1b-nsp14 | COVID-19 | Hong Kong | TGG GGY TTT ACR GGT AAC CT | AAC RCG CTT AAC AAA GCA CTC | TAG TTG TGA TGC WAT CAT GAC TAG | (40) |
| RdRp/Orf1 | COVID-19 | Germany | GTG ARA TGG TCA TGT GTG GCG G | CAR ATG TTA AAS ACA CTA TTA GCA TA | CAG GTG GAA CCT CAT CAG GAG ATG C | (19) |
| RdRp/Orf1 | COVID-19 | China | CCC TGT GGG TTT TAC ACT TAA | ACG ATT GTG CAT CAG CTG A | CCG TCT GCG GTA TGT GGA AAG GTT ATG G | (41) |
| orf1a | COVID-19 | China | AGAAGATTGGTTAGATGATGATAGT | TTCCATCTCTAATTGAGGTTGAACC | TCCTCACTGCCGTCTTGTTGACCA | (42) |
The Common Primers Used for the Detection of SARS-CoV-2
9. Detection of SAR-CoV-2 RNA Using RT-PCR
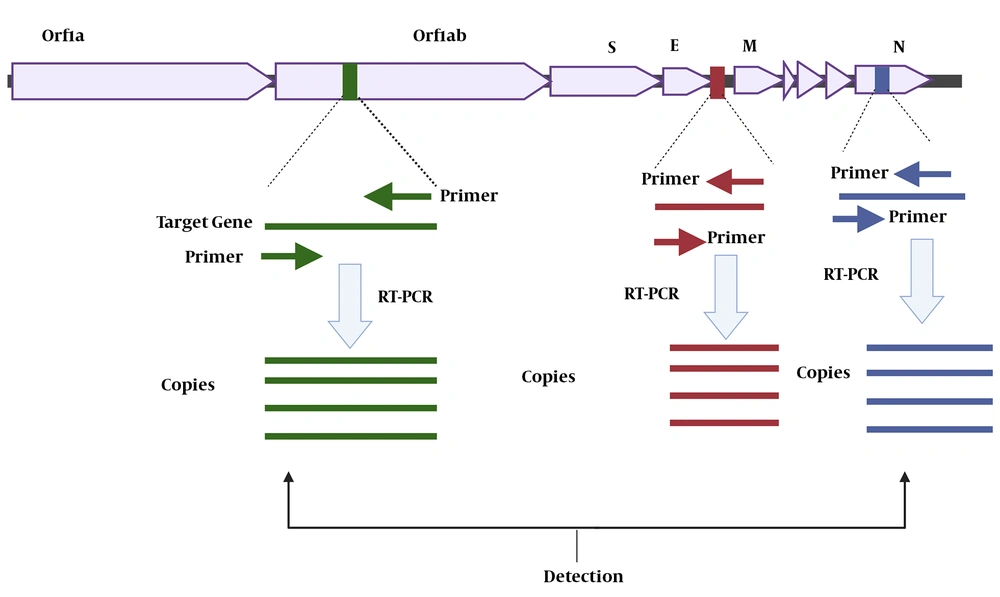
Currently, the identification of viral RNA by RT-PCR is regarded as the gold standard for COVID-19 diagnosis and treatment follow-up (61). The detection of this pathogenic virus in respiratory specimens requires isolating the genetic material of the virus (RNA) followed by its conversion to cDNA and finally cDNA amplification. In this process, specific primers (forward and reverse) are used to amplify the target gene(s). The two most common targets are the ORF1ab and "N" genes, both of which are positive reference genes (61, 62) (Figure 3). The amplification condition can be set as follows: 50°C for 15 min, 95°C for 3 min, and then 45 cycles of 95°C for 15 s and 60°C for 30 seconds (59, 62). Usually, fluorescence acquisition in qPCR with dsDNA binding dyes occurs during the melting process at a temperature between the melting points of primers and the amplicon (63). The fluorophore-quencher probe is cleaved during thermocycler senses the fluorescent signal and tracks amplification progression in real-time (64). The diagnostic algorithm used to detect SARS-CoV-2, the causative agent of COVID-9, uses an initial screening to detect the presence of SARS-CoV-1, SARS-CoV-2, and other viruses belonging to the Sarbecovirus subgenus. For confirming the diagnosis, probes, and primers unique to the RdRP gene (RNA-dependent polymerase RNA) are used. Many labs in the world use the diagnostic algorithm and reagents employed by Corman et al. (37) in Cuba, as well as their interpretation to detect SARS-CoV-2 (37). For the "E" gene, a sample is considered positive if Ct is ≤ 36. In these cases, the diagnosis is confirmed using the RdRP gene, whose positivity is warranted at a Ct of ≤ 40. According to a report from a hospital in Wuhan, China, positive results were obtained for a patient only when both target genes (ORF1ab and N) were assessed (62). Accordingly, the result was considered positive when the Ct value was < 37 and negative when Ct was ≥ 40. A moderate viral load, defined as a Ct value from 37 to < 40, needed re-testing for confirmation (61). For widespread use of these molecular technologies in armed forces, military scientists must focus on designing a portable RT-PCR-based technology that is capable of integrating all procedures in one device and detecting infectious agents at affordable costs.
Figure 3 shows the target genes (Orf1ab, "E", and "N") used for the diagnosis of COVID-19 applying specific forward and reverse primers. These genes are then amplified via RT-PCR to produce a million copies of them. The presence of the genes generates signals that are detected by the RT-PCR device, confirming the diagnosis of COVID-19.
10. Conclusions
The achievements acquired by scientists in the fields of diagnosis and treatment of COVID-19 since the beginning of the outbreak encompass the sequencing of the viral genome, development of effective and reliable methods for detecting the virus, drug repurposing, and producing vaccines, which are currently on trials. The global spread of COVID-19 has exposed military personnel to all sorts of threats, and therefore, there is a need to secure military organizations by implementing disease surveillance programs and the effective diagnosis and rapid management of the disease using molecular tools such RT-PCR. Currently, RT-PCR is regarded as the gold standard for detecting COVID-19 in respiratory samples, including bronchoalveolar lavage fluid, sputum, nasal and pharyngeal swabs, fibrobronchoscope brush biopsy, as well as in feces, and blood. Reverse-transcriptase-PCR procedures ensure the early detection and rapid and effective control of the disease. The method is sensitive and reliable as it detects the genetic materials of pathogens. In some diseases where antibodies are initially detected using other methods, for example, in HIV, RT-PCR is employed to confirm the presence of the viral genetic material, making the diagnostic procedure more efficient. In conclusion, RT-PCR will continue to remain a reliable diagnostic test for detecting SARS-CoV-2 and monitoring patients in the course of treatment.