1. Background
Pseudomonas aeruginosa is one of the opportunistic pathogens causing hospital-acquired infections, especially in some groups of patients such as immunocompromised, burns, and cystic fibrosis (1). Infections causing multidrug-resistant P. aeruginosa isolates increase morbidity and mortality, enhance treatment costs, and prolong hospitalization period (2).
Carbapenems are the drug of choice in treating multi-drug P. aeruginosa infections. The prevalence of carbapenem resistance among P. aeruginosa has increased worldwide (3). Because of some intrinsic resistance to antibiotics, the treatment of pseudomonas infections is a challenge. The main mechanisms for the high rates of antibiotic resistance are decreased drug accumulation because of the low permeability of cell wall and efflux pumps, chromosomal mutations, and the transfer of resistance genes by plasmids, transposones, and bacteriophage (4).
Among the carbapenemase types in Pseudomonas spp, according to the molecular classification of Ambler, there are serine β-lactamases such as KPC (Klebsiella pneumoniae carbapenemases) and GES (Guiana extended-spectrum) in Class A and OXA-198 (Oxacillinases-198) in Class D (5, 6). New metallo-beta-lactamase (MBL) enzymes, which may be responsible for the growing carbapenem resistance of non-fermentative Gram-negative (NFGN) bacilli, have been identified in recent years, which are spread worldwide (7). Many carbapenemases have been identified in Pseudomonas species and encompass metallo-β-lactamases (MBL) in Class B, including imipenemase (IMP), Verona integron-mediated metallo-β-lactamase (VIM), Sao Paulo MBL (SPM), Seul imipenemase (SIM), Australian imipenemase (AIM), German imipenemase (GIM), Dutch imipenemase (DIM), and new Delhi metallo β-lactamase (NDM) (8, 9).
2. Objectives
This study aimed to investigate the presence of the VIM, NDM, IMP, GES, SPM, GIM, and SIM genes accounting for the increase of resistance to carbapenem antibiotics in P. aeruginosa isolates.
3. Methods
In this study, 200 carbapenem-resistant P. aeruginosa isolates that isolated from different clinical samples were tested. Conventional methods (Gram staining, oxidase test) and the Vitek-MS (Biomeirux, France) automated system were used to identify the isolates. The antibiotic susceptibility was also tested using the Vitek 2 Compact (Biomeirux, France) automatization system. The P. aeruginosa isolates were stored at -20°C for further molecular studies. The presence of IMP, VIM, NDM, GES, SPM, GIM, and SIM genes was investigated by the multiplex PCR. Table 1 presents the primers used in the study (10-12). The boiling method was also used for the DNA extraction.
| Genes | Primers Sequence | Expected Amplicon Size (bp) | References |
|---|---|---|---|
| VIM | 389 | (10) | |
| VIM-F | GTT TGG TCG CAT ATC GCA AC | ||
| VIM-R | AAT GCG CAG CAC CAG GAT AG | ||
| NDM | |||
| NDM-F | GCA GCT TGT CGG CCA TGC GGG C | 782 | (10) |
| NDM-R | GGT CGC GAA GCT GAG CAC CGC AT | ||
| IMP | 188 | (12) | |
| IMP-F | GGA ATA GAG TGG CTT AAT TCT C | ||
| IMP-R | CCA AAC CAC TAC GTT ATC T | ||
| GES | 863 | (11) | |
| GES-F | ATG CGC TTC ATT CAC GCA C | ||
| GES-R | CTA TTT GTC CGT GCT CAG GA | ||
| SPM | 271 | (12) | |
| SPM-F | AAA ATC TGG GTA CGC AAA CG | ||
| SPM-R | ACA TTA TCC GCT GGA ACA GG | ||
| GIM | 477 | (12) | |
| GIM-F | TCG ACA CAC CTT GGT CTG AA | ||
| GIM-R | AAC TTC CAA CTT TGC CAT GC | ||
| SIM | 570 | (12) | |
| SIM-F | TAC AAG GGA TTC GGC ATC G | ||
| SIM-R | TAA TGG CCT GTT CCC ATG TG |
Sequence of Primers in the Study
4. Results
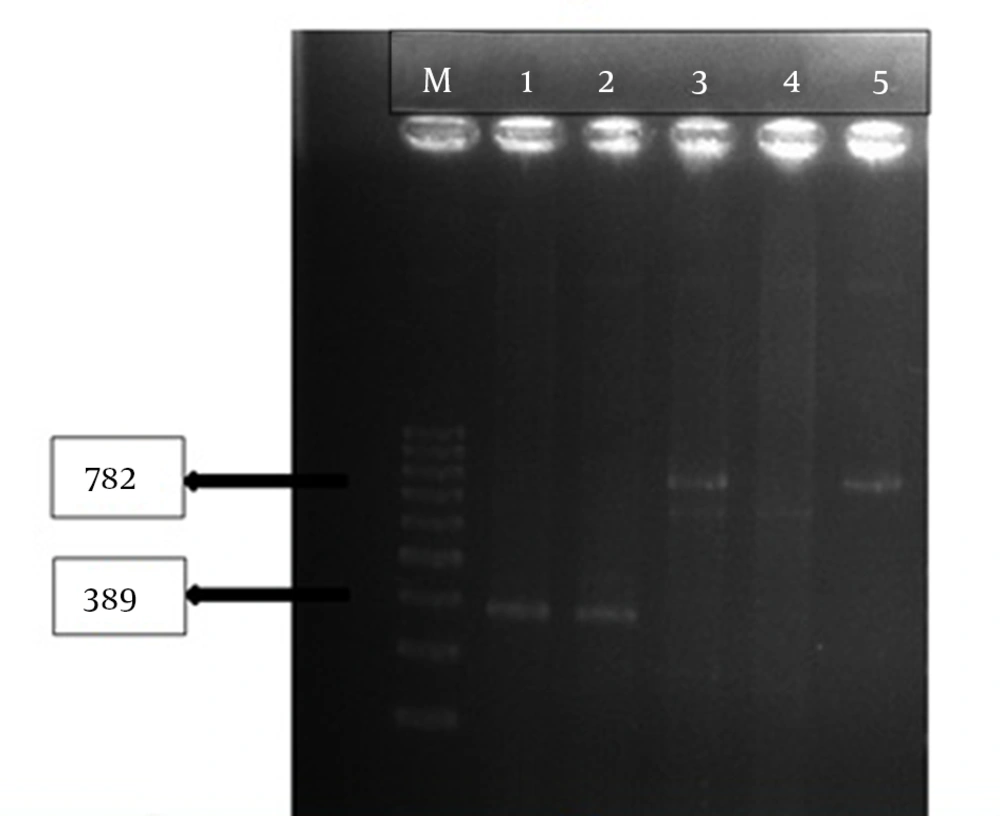
This results indicated that tracheal aspirate cultures were the most common samples (34.5%) of the carbapenem-resistant P. aeruginosa isolates (Table 2). The VIM and NDM genes were noticed in two of the P. aeruginosa (P32, P190) isolates (Figure 1).
| Specimens | No. (%) |
|---|---|
| Tracheal aspirate culture | 69 (34.5) |
| Sputum culture | 36 (18) |
| Urine culture | 34 (17) |
| Exudate culture | 22 (11) |
| Blood culture (Aerob) | 15 (7.5) |
| Wound culture | 10 (5) |
| Catheter | 5 (2.5) |
| Sterile body liquid | 5 (2.5) |
| BOS | 2 (1) |
| Swab | 1 (0.5) |
| Bronchoalveolar lavage (BAL) culture | 1 (0.5) |
Distribution of Specimens of Pseudomonas aeruginosa Isolates
5. Discussion
Carbapenems are among the best options for treating infections caused by multidrug-resistant Gram-negative bacteria; however, their inappropriate usage has increased resistance. Due to the increased prevalence of the MDR strains, the treatment of infections caused by P. aeruginosa is challenging (13, 14). The “Carbapenem Antimicrobials Pseudomonas Isolate Testing at regional Locations (CAPITAL) surveillance programme” in 2010 indicated that the overall rates of carbapenem-resistant P. aeruginosa ranged from 7.4 to 35.4%. Moreover, the rates of carbapenem resistance in a meta-analysis study in 2015 ranged from 8.7 to 50.4% among the P. aeruginosa isolates (15, 16). In this regard, the upregulation of efflux pumps, decreased outer membrane permeability, and acquired metallo-beta-lactamases (MBL) are introduced as the main factors leading to carbapenem resistance (17). The P. aeruginosa MBLs were detected in the early 1990s, the first representatives of which are IMP and VIM type enzymes (18, 19).
In Brazil, the SPM-1 gene, the most frequent metallo-lactamase gene, was first introduced in 2002 and then reported in different sites (20, 21). The SPM-1 gene region was detected in 33 out of 129 carbapenem-resistant P. aeruginosa isolates isolated from the hospitalized patients during 1998 - 2012, in four and three of which the VIM-2 and GES-3 genes were detected, respectively. Furthermore, the SPM-1 and KPC-2 links were found in the nine strains, and the SPM-1, VIM-2, and KPC-2 link was observed in one strain (22).
Carbapenem resistance genes (VIM, PER, IMP, GES, KPC, OXA) were examined in the ceftazidime-resistant P. aeruginosa strains (n = 195) isolated from the hospitalized patients using the PCR test. In this study, the OXA-10 (n = 5), OXA-14 (n = 4), VIM-2 (n = 4), IMP-1 (n = 2), and GES-1 (n = 26) determinants were detected; however, all isolates were negative for the PER and KPC genes (1).
Amoureux et al. isolated IMP-19 producing P. aeruginosa from seven patients with nosocomial infections linked to contaminated sinks in France (23). Their findings showed that the prevalence of MBL producers among imipenem-resistant P. aeruginosa was lower in France than the other countries, and that the VIM producers were mainly determined during the outbreaks (8, 24-26). Various MBLs, including IMP, VIM, NDM, SPM, GIM, and SIM were identified worldwide; however, NDM, VIM, and IMP genes are frequently observed in P. aeruginosa in India (27). The IMP and VIM-producing Pseudomonas isolates have been reported in different geographical regions (28).
Hakemi Vala et al. evaluated the presence of classes A, B, and D (IMP, VIM, SPM, KPC, GIM, DIM, BIC, OXA-48, CTX-M15, and NDM genes) β-lactamases among P. aeruginosa isolates (29). They reported that the frequencies of the positivity of the CTX-M15 and IMP genes among 47 P. aeruginosa isolates were four (8.5%) and one (2.1%), respectively (29). Fallah et al. showed that the P. aeruginosa isolates harboured the IMP gene in their study (30).
Zafer et al. examined the prevalence of metallo-β-lactamases (MBL) in P. aeruginosa isolates (n = 122) collected from two different hospitals in Cairo, Egypt, and found out that the VIM-2 gene was the most prevalent MBL gene (31). In a study on the presence of the IMP-6, VIM-2, KPC, GES, NDM, and OXA-48 genes, IMP-6 and VIM-2 MBLs were identified in 17 and four of the 329 P. aeruginosa isolates (32).
VIM-2 is the dominant MBL gene associated with the outbreaks caused by MBL producing P. aeruginosa worldwide (33, 34).
Resistance in P. aeruginosa is an emerging problem worldwide, and carbapenem resistance is one of the critical problems caused by the spread of carbapenemases. VIM is frequently isolated from carbapenem-resistant P. aeruginosa isolates. In the present study, we detected one isolate harbouring VIM. Moreover, NDM was isolated from the carbapenem-resistant isolates during the last decade, and one isolate was positive for NDM in our study.