1. Background
Pseudomonas aeruginosa (P. aeruginosa), rod-shaped gram-negative bacteria, can cause acute infections in humans (1). These bacteria are multidrug resistant pathogens recognized for their ubiquity, their intrinsically advanced antibiotic resistance mechanisms and association with serious infections in hospitalized patients, such as ventilator-associated pneumonia and various sepsis syndromes (1-3).
Pseudomonas aeruginosa produces a broad spectrum of virulence factors, the expression levels of which are firmly adjusted (4). The key to this adjustment is quorum sensing (QS), a ubiquitous cell density-dependent cell-to-cell communication system (4-6).In P. aeruginosa, similar to other bacteria, QS controls formation of biofilm, virulence factors secretion and DNA exchange (7-9). It has been shown that the growth of biofilm is the main bacterial property and plays a critical role in infections (10, 11).
On the other hand, numerous infections require the formation of bacterial biofilms, which are communities of bacterial that proliferate and fix on surfaces and are hidden by exopolymers (12).
It must be noted that after formation, it is difficult to eradicate the biofilm and this becomes a principle of secondary infection (4, 13).
Furthermore, bacteria hidden in biofilms are more resistant against antibiotics. Therefore, in this condition, treatment of infections often fails (14, 15).
Gram-negative bacteria, such as P. aeruginosa, produce and release N-acyl Homoserine Lactones (AHLs) by QS. Suitable concentrations of AHLs then bind to specific receptors and make dimers or polymers of the activated receptor-AHL complex which, in turn, acts as transcriptional modifiers of target genes in the QS region (16, 17).
Hence, the expression of virulence genes and induction of biofilm formation, through a cascade of regulatory events, is coordinated by AHLs (4, 18). Therefore, the QS system plays a crucial role in organizing infections and enhancing resistance to antibiotics (6, 15).
Discovery and development of drugs that inhibit the QS system cause a major innovation in the eradication of antibiotic resistant bacteria (19, 20). It has been found that some QS Inhibitors (QSIs) like patulin, a mycotoxin produced by a variety of molds, make P. aeruginosa more susceptible to tobramycin, an aminoglycoside antibiotic (21-23).
It must not be forgotten that P. aeruginosa cell-to-cell signaling is not limited to AHLs. Two distinct, yet related QS circuits, have been identified in P. aeruginosa. Both of these systems are genetically similar in that they consist of genes encoding transcriptional activator proteins (lasR and rhlR) as well as genes responsible for the production of AHL signaling molecules (lasI and rhlI) (24, 25).
In the QS systems, the intercellular signals for the las and rhl are N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL) and N-(butanoyl)-L-homoserine lactone (C4-HSL) (26). It has been shown that these signals regulate hundreds of genes, indicating 4% to 12% of the P. aeruginosa genome (27, 28).
Furthermore, 3-oxo-C12-HSL and C4-HSL, 2 bacterial cell-to-cell signals, are essential for the production of various extracellular virulence factors and the development of a differentiated biofilm in P. aeruginosa (29, 30).
It has been reported that P. aeruginosa produces another signal molecule, 2-heptyl-3-hydroxy-4-quinolone, which has been determined as the pseudomonas quinolone signal (PQS), a unique cell-to-cell signal (30, 31). Furthermore, PQS was shown to control the expression of lasB, which encodes for LasB elastase, a major virulence factor (32, 33). Moreover, the P. aeruginosa las and rhl QS systems were interfering in the synthesis and bioactivity of PQS, respectively (31).
Transcriptome analysis studies have shown that PQS directly and indirectly activates 92 and 143 genes, respectively (34, 35).
For this purpose, PQS binds to and activates PqsR (a LysR-type transcriptional regulator), and subsequently induces the expression of the pqsABCDE operon (36, 37). The pqsABCD genes of this operon are required for PQS synthesis, yet, the function of the pqsE gene is not clear (38-41). It has been reported that the pqsE gene is required for the production of several PQS-controlled virulence factors (42). Furthermore, it has been suggested that the regulatory activity of PqsE is unrelated to PqsR and PQS, yet, related to the rhl QS system (31, 42).
2. Objectives
This study aimed at identifying putative inhibitors of LasR by structure-based virtual screening on ZINC database, testing prospective candidates on pqsE with computer aided inhibition studies followed by structure analysis, and to determine the extension of specific drugs with potential anti-virulence properties for the treatment of infections induced by drug-resistant P. aeruginosa.
3. Methods
AutoDock Vina is a program for molecular docking and virtual screening that uses a command line interface and only accepts 1 receptor and ligand in each run. As structural virtual screening is a repetition of docking cycles for thousands of ligands, these limitations make it a time consuming task. Therefore, a software was developed using Microsoft visual studio 2012 (Visual Basic) to automate virtual screening steps. The program was designed to accept the database and receptor files through graphical user interface and to extract and convert the database to single ligands and then to run the docking cycle for each ligand with the same receptor. The program was set to output a list of ligands sorted by descending affinity in conjugation with their respective docking energy (43).
Autodock Vina achieves an approximately 2 order of magnitude speed-up compared with AutoDock 4, a molecular docking software. In addition, it significantly improves the accuracy of the binding mode predictions, judging by the tests on the training set used in AutoDock 4 development. Furthermore, speed-up is achieved from parallelism, by using multithreading on multicore machines. AutoDock Vina automatically calculates the grid maps and clusters the results in a way transparent to the user (43, 44).
The ZINC database was downloaded from its website. The 3D structures of LasR (PDB ID: 2UV0) and pqsE (PDB ID: 2Q0J) were downloaded from RCSB protein data bank and were used as receptors. To reduce the time needed for screening and to eliminate further need for animal and human safety tests, the zinc database was filtered so that only Food and Drug Administration (FDA) approved compounds were taken into account. About 2300 compounds were then used for virtual screening and subsequent docking studies.
First screening was performed on LasR. Autodock tools were used to prepare the receptor by adding partial Kollman charges and missing polar hydrogen. The region of interest to be used by Autodock vina was defined as the center of the ligand already present in the crystal structure; the ligand and other unnecessary molecules were removed before commencement of the docking process. The size of the grid was set to 17, 17, and 23 angstroms in x, y, and z directions. Each compound was subjected to 10 runs of Vina with exhaustiveness value set to 8 and selection of LasR as the receptor. The same set of ligands was then docked into pqsE, following the same procedure. The size of the docking grid was set to 17 in all 3 dimensions. Ligands were sorted based on descending affinity to each receptor, and top 50 substrates were selected giving the same weight to LasR and pqsE binding affinity.
Ligands were finally inspected by the ViewerLite program for placement accuracy and formation of proper hydrogen bonds and van der Waals interactions.
4. Results
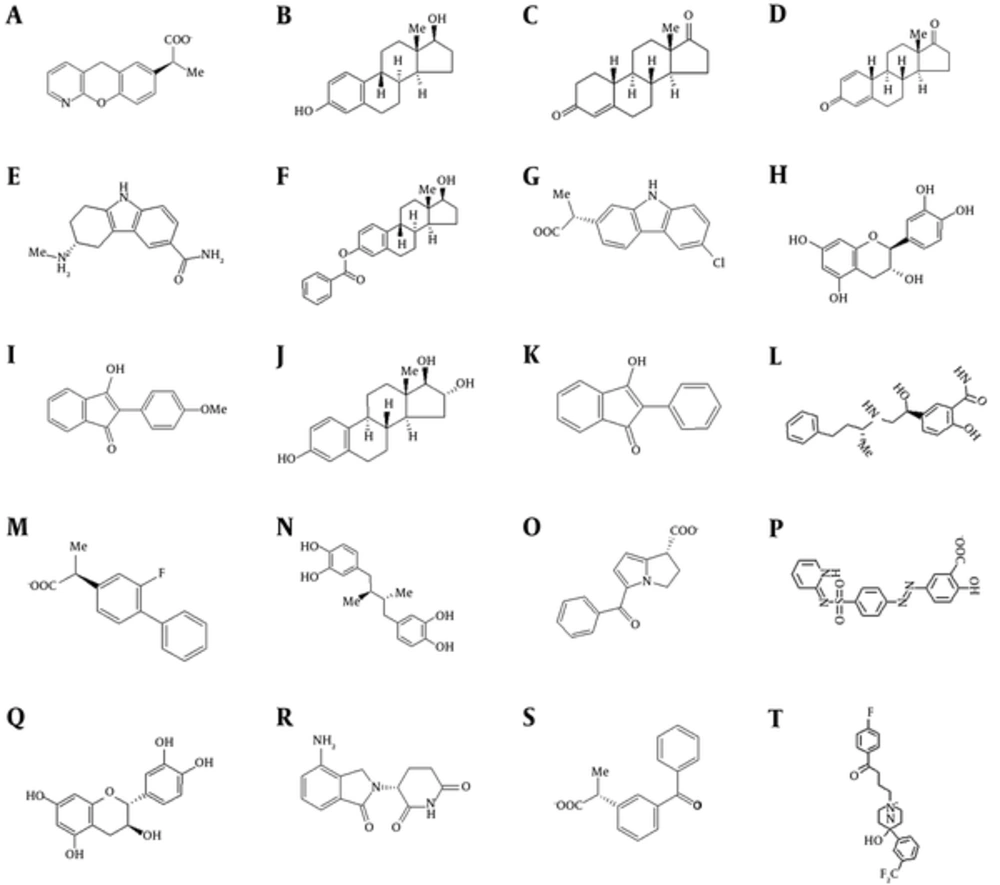
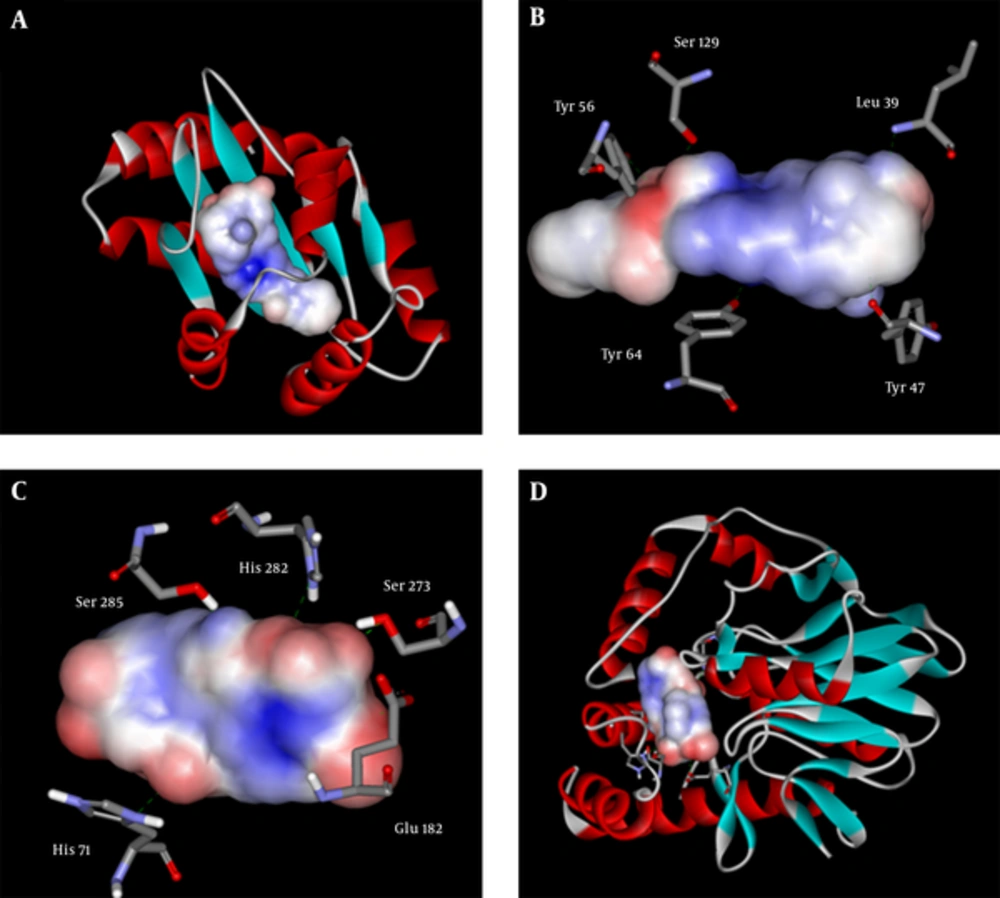
Top structures and their affinity to the 2 receptors are detailed in Table 1 and Figure 1. The data analysis showed that these compounds are very similar in structure. These inhibitors can mainly be categorized in 2 major groups, steroids and some subfamilies of non-steroidal anti-inflammatory drugs (NSAIDs). It can be inferred that having 2 polar groups with a distance similar to oxygen to oxygen distance between 3- and 17-hydroxyl groups of estradiol is required for proper interaction with the active site. These 2 groups are required for interaction with Tyr 47 and Ser 129 in LasR. For PqsE, they interact with Ser 273 and His 71. These interactions are depicted in Figure 2B and 2C. It seems that although the presence of these 2 groups is essential for proper binding, the distance is somewhat flexible, and ligands without fused rings with variation in distance also bind with lower affinity.
A, Pranoprofen; B, Estradiol; C, 19-Norandrostendione; D, Estrone; E, Triptane; F, Estradiol Benzoate; G, Carprofen; H, Catechin; I, Anisindione; J, Estriol; K, Phenindione; L, Labetelol; M, Flubiprofen; N, Nordihydroguaiaretic acid; O, Ketorolac; P, Sulfasalazine; Q, Cianidanol; R, Lenalidomide; S, Ketoprofen; T, Fluroperidol.
| Structure Name | LasR | pqsE |
|---|---|---|
| Docking energy | ||
| Pranoprofen | -9.3 | -10.4 |
| Estradiol | -8.8 | -10.4 |
| 19-Norandrostendione | -10 | -10.2 |
| Estrone | -9.3 | -10.3 |
| Triptane | -9 | -10.3 |
| Estradiol Benzoate | -8.7 | -12.8 |
| Carprofen | -9.8 | -9.6 |
| Catechin | -8.1 | -10.3 |
| Anisindione | -10.2 | -8.1 |
| Estriol | -10.0 | -8.6 |
| Phenindione | -9.7 | -9.0 |
| Labetalol | -10.2 | -8.0 |
| Flurbiprofen | -9.7 | -9.1 |
| Nordihydroguaiaretic acid | -8.0 | -10.1 |
| Ketorolac | -9.7 | -8.5 |
| Sulfasalazine | -10.5 | -7.5 |
| Cianidanol | -10.3 | -7.6 |
| Lenalidomide | -9.6 | -8.6 |
| Ketoprofen | -9.5 | -9.4 |
| Fluroperidol | -11.3 | -7.3 |
aStructure of the compounds are presented as Figure 1.
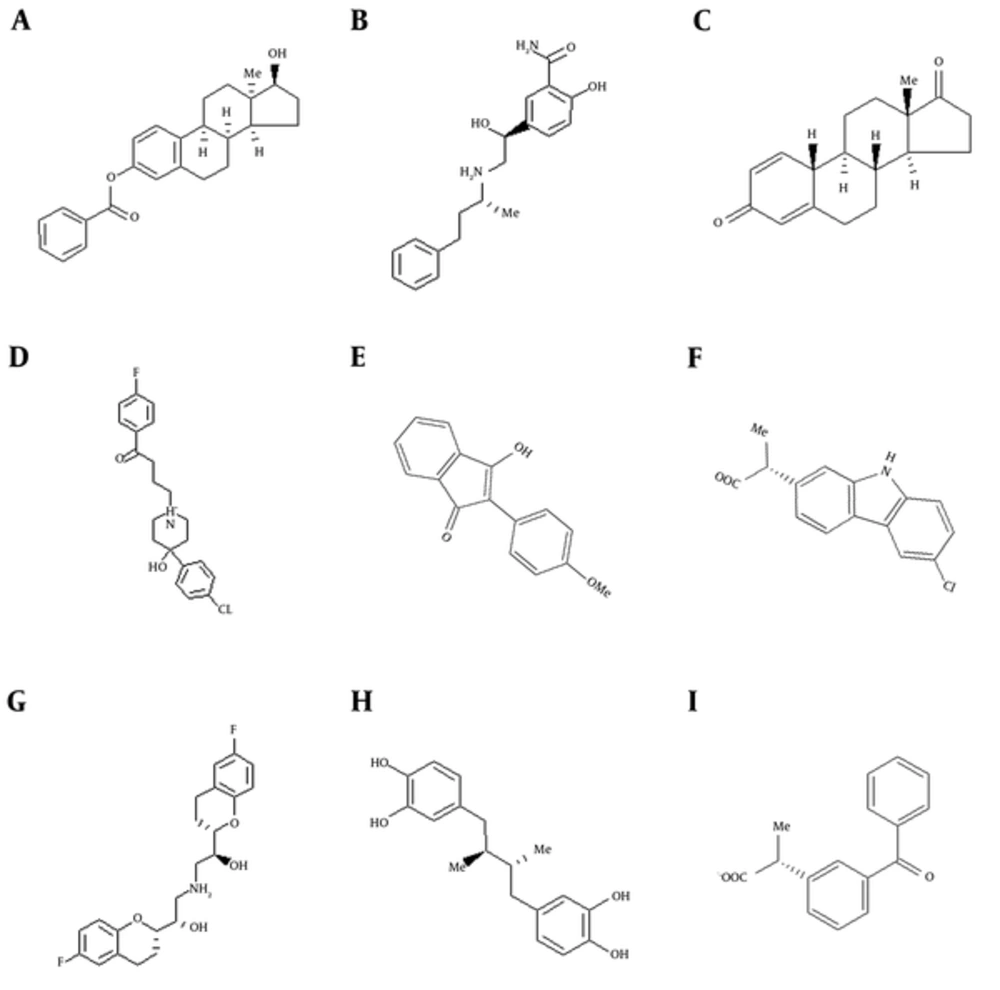
Figure 2A and 2D show the 2 proteins with surfaced ligands inside. It can be observed that due to smaller size of pqsE’s active site, compounds with higher affinity to the mentioned protein have smaller sizes in comparison to LasR inhibitors. Figure 3 demonstrates the difference between compounds with more affinity to pqsE or LasR. The LasR inhibitors are very similar to pqsE inhibitors on one end and on the other end it seems that having only an aromatic ring is sufficient for proper attachment. The mentioned ring forms aromatic interactions with phenol rings of Tyr 56 and Tyr 64. Having an electron substituted on this ring ameliorates binding affinity by facilitating formation of hydrogen bonds.
High similarity was seen between some of compounds listed in Table 1 and Figure 1 with tryptophan. Aromatic amino acids are known to be inhibitors of the pathway by regulating the first step of chorismate biosynthesis in the shikimate pathway (45). It can also be concluded from the data in Figure 3 that bigger compounds with a good degree of flexibility can fit the smaller active site of pqsE with partially degraded affinity.
5. Discussion
Pseudomonas aeruginosa is a multi-drug resistant pathogen known for its ubiquity, especially in war zone hospitals. It is well known that this opportunistic pathogen has intrinsically advanced antibiotic resistance mechanisms via coordinate gene expression and produces serious infections (1-3).
For these bacteria, quorum sensing (QS) plays an essential role for competitiveness in clinical or environmental sites (21). Bacterial QS system inhibition by QS Inhibitors (QSIs) may be applied in various fields such as medicine, food technology, and agriculture (4, 22).
Discovery and development of QSIs is attractive because these drugs reduce bacterial pathogenicity, via inhibition of QS and anti-biofilm effects, and so indirectly inhibit bacterial growth (13, 22). Furthermore, these drugs are not directly involved in the development of antibiotic resistance mechanisms (23).
For this reason, this study attempted to find novel targets with bacterial QS inhibition and anti-biofilm properties.
It is known that a broad range of P. aeruginosa virulence-related factors are regulated by QS LasR transcriptional regulator (30). Furthermore, although pqsE gene is not required for the synthesis of PQS, yet, is required for the production of many PQS-controlled virulence factors and for virulence in several models of infection. In other words, PqsE, as a regulator, can activate PQS-controlled genes without presence of PqsR and PQS (42).
Thus, it was concluded that dual inhibitors of LasR and pqsE would be of uttermost efficacy.
In this investigation, the above-mentioned pqseE was selected. On the other hand, the 3D structures of transcriptional regulators from P. aeruginosa involved in QS was elucidated (46, 47).
Because virtual screening is an integral part of many drug-discovery efforts and is widely used in pharmaceutical research, we applied this technique in a search for novel QSIs.
Considering that the currently available QSIs are inappropriate for human use, there is special interest in discovery of novel chemical agents. According to this, virtual screening approaches involve database filtering to exclude compounds containing toxic, reactive, or otherwise undesired groups (48).
In the present study, a database of 2300 compounds was screened because they were already FDA-approved for human use. Among the 20 introduced compounds, members of different drug groups can be found. The major sub group of the compounds is from the non-steroid anti-inflammatory drug group. It can be inferred that the active site of these proteins somewhat resembles COX. Steroids are also prominent among the suggested compounds. Thus, as a general guideline, bi-therapy of antibiotics and anti-inflammatory drugs of any type may reduce bacterial resistance in P. aeruginosa infections.
The software designed in this study provided a graphical user interface to Vina. It easily inputs required docking parameters using textboxes and buttons that reduce user training time in comparison to command-line interface of Vina. There is also no need for conversion of ligand files by the user. It detects ligand file formats and uses OpenBabel (49) to convert ligands to the PDB format. It provides Autodock vina for each ligand of a given dataset with parameters set in its GUI. After completing docking cycles, the software runs vina split to split docking results and analyzes the data. Finally it collects the best binding energy for each compound from the result files and displays and saves them in a text file in ascending order, assigning a rank number to each compound.
5.1. Conclusions
The present study provides new means of inhibiting bacterial resistance without relying on more potent bactericides or bacteriostatic compounds. The necessity to develop new anti-pathogenic drugs is critical, especially in the case of infections induced by P. aeruginosa, as it is a multidrug resistant pathogen and has intrinsically developed antibiotic resistance mechanisms by QS system and biofilm formation (50).
Therefore, compounds characterized in the present study prepare the potential for antibiotics to target infections that are insistently difficult to battle with the current antibiotics.