1. Background
Virtual screening (VS), as a computational technique, is being used widely by pharmacologists and medicinal chemists in drug discovery research. By using computers, it deals with the quick search of large libraries of chemical structures, to identify those structures, which are most likely to bind to a drug target, typically a protein receptor or enzyme (1-3).
The VS is defined as “automatically evaluating very large libraries of compounds using computer programs” (4). As this definition suggests, VS has been largely focusing on questions like how can the enormous chemical space of 1060 conceivable compounds (5) be filtered to a manageable number that can be synthesized, purchased and tested. Although filtering the entire chemical universe might be a fascinating question, more practical VS scenarios focus on designing and optimizing targeted combinatorial libraries and enriching libraries of available compounds, from in-house compound repositories or vendor offerings.
Two major strategies are used in VS: 1) structure based VS method ,which is employed when three dimensional (3D) structures of the targets are available and 2) ligand based VS that is used where 3D target structures are unknown (6). Structure based VS is dependent on the knowledge of target structure. A ligand collection is tested on the target by “docking” and a quantified interaction score is used to identify candidate lead compounds. Therefore, structure based VS is independent on the existence of known active lead compounds (7). Another approach to ligand-based VS is to use two-dimensional (2D) chemical similarity analysis methods (8) to scan a database of molecules against one or more active ligand structures. Another method of ligand-based VS is based on searching molecules with shapes similar to those of known active sites, as such molecules will fit the target’s binding site and, hence, will be likely to bind the target. A number of prospective applications of this class of techniques have been identified in the literature (9, 10).
Docking is a method which predicts the preferred orientation of one molecule to a second, when bound to each other to form a stable complex (11). Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules, using, for example, scoring functions. Docking is frequently used to predict the binding orientation of small molecule drug candidates to their protein targets to, in turn, predict the affinity and activity of the small molecule. Therefore, docking plays an important role in the rational drug design (7).
Various docking software have been developed for virtual screening, since the initiation of University of California, San Francisco dock (12, 13), such as GOLD (14), GLIDE (15) or AutoDock Vina (16). Multiple researchers have reported successful identification of lead compounds, using docking based VS methods (17). Ligand based VS is based on the assumption that structurally similar compounds are likely to exhibit similar activities. Structural, physiochemical and energetic properties are used when screening large databases for related or novel chemical compounds (18). The pharmacophore-based database searching technique is a widely used VS strategy (19), which relies on knowledge of the biological activity of multiple hits, when identifying key features, during a search. A pharmacophore is a spatial arrangement of features that allows a compound to interact with a target receptor and generate a response. In research,, both methods can be used together or separately.
A very widely used data base for virtual screening is “ZINC” (it is not commercial) (20). It is a curate collection of commercially available chemical compounds, prepared especially for VS. ZINC differs from other chemical databases, because it aims to represent the biologically relevant, 3D structure of the molecule. This database is updated regularly and may be downloaded and used, free of charge. The latest release of the website interface is “ZINC 12” (2012). The database contents are continuously updated, while static subsets are generated regularly and are dated.
Visual screening tools in hand are usually command-line oriented and lack an intuitive graphic user interface (GUI) and this has prevented the main audience of such software, which are medicinal chemists, pharmacologists and other pharmaceutical scientists, with mainly novice computer skills to actively engage.
Because there is a growing need for easy to use software, in this field, in this study, we have developed a VS piece of software: IranVScreen, which is an easily operable tool for medicinal chemists and pharmacologists, to carry out multiple practical virtual screening tasks, while also providing the facility to extract the results. IranVScreen integrates several static subsets of ZINC database, which further facilitates the task for the researcher and allows for very fast reclamation of results. IranVScreen provides a very intuitive all-in-one GUI to carry out multiple virtual screen tasks, with several mouse clicks, with minimal requirement of skill.
2. Objectives
The main objectives were to provide a very easy to use software, with comprehensive GUI, and to integrate several databases to minimize the need of computer knowledge for the users and to provide them with an all-in-one solution to VS.
3. Materials and Methods
The software was developed using Microsoft’s Visual Studio 2012. The main application was developed in Visual basic. The GUI was developed as a windows forming application on .Net framework 4.5. The codes for controlling the integrated software and interpretation of the results were written in C++. IranVScreen integrates the programs detailed in the following sections and provides a uniformed interface to the user.
3.1. Open Babel
Open Babel is a free software, a chemical expert system mainly used for converting chemical file formats (21). Due to the strong relationship to informatics, this program belongs more to the category cheminformatics than to molecular modeling. It is available for Windows, UNIX and Mac OS. It is distributed under the GNU General Public License. Open Babel includes two components, a command-line utility and a C++ library. The command-line utility is intended to be used as a replacement for the original Babel program, to translate between various chemical file formats. The C++ library includes all of the file-translation codes, as well as a wide variety of utilities to foster development of other open source scientific software.
3.2. AutoDock Vina
AutoDock Vina is a new program for molecular docking and VS. It achieves an approximately two orders of magnitude speed-up, compared with the molecular docking software previously developed in the same lab (AutoDock 4), while also significantly improving the accuracy of the binding mode predictions, judging by the tests on the training set used in AutoDock 4 development. Further speed-up is achieved from parallelism, by using multithreading on multicore machines. AutoDock Vina automatically calculates the grid maps and clusters the results, in a way transparent to the user (16).
3.3. Integration
Open Babel provides an intuitive application program interface (API) and related documentation. An API is a set of functions, routines and protocols used to build computer software. The APIs are like building blocks of software, as every programmer can put useful ones together and build new programs. The provided classes were used to instantiate required objects. Vina integration was done by including and calling suitable dynamic link library files. Several useful ZINC subsets were also compressed and included to ease the process of database retrieval.
4. Results
4.1. Interface and Features
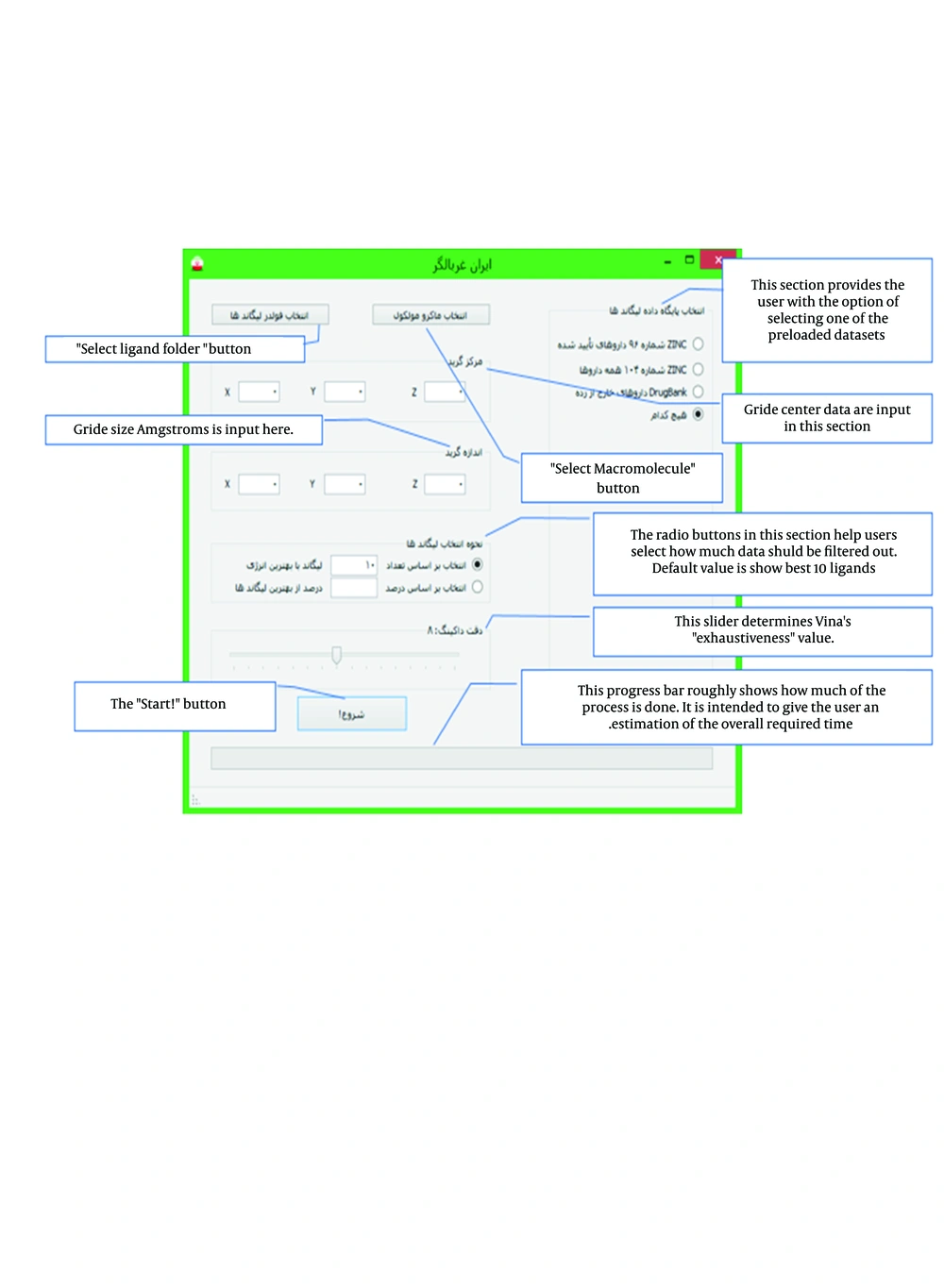
As shown in Figure 1, on the right pane, radio buttons are provided for the user to choose from one of the preloaded ZINC subsets. Whether or not the user decides to use one of preloaded databases, any ligand placed in “Ligand Folder” selected by “Choose Ligand Folder” button will be added to the selection.
Also, on the top of the left pane, the user is provided with two buttons: “Select Macromolecule” and “Select Ligand Folder”. Clicking the first button displays a browse for file dialog box to select the macromolecule used for VS. The second button shows a browse for folder dialog box to select a folder that contains ligands to be screened. Even if there are no additional ligands, this folder should be selected since this folder is used for storage of temporary files and results.
Under the mentioned buttons, there are six text boxes to receive grid size and center. These values must be determined using other tools, such as SPDBV or MGL tools. The values should be entered in Angstroms (Å).
There is another section below the said boxes by which the user can choose how many compounds are desired to be saved. Two options are provided: 1) the user can enter the exact number of required ligands. In this case, the final results file contains this number of best results sorted from best to worst. 2) The user can input the percentage of ligands to be saved. If this option is selected, the number is calculated by multiplication of the entered percentage, by the total number of ligands.
Underneath this section, a slider is placed for the user, to adjust the desired quality of docking. The placement of the slider determines the exhaustiveness factor of Vina, between 0 and 16.
The start button, placed in the bottom left section, triggers the virtual screening process. First, the form is validated to see if the required file and folder are selected. If any dataset is selected, the sdf file is copied to the working directory. The folder is then inspected and a list of molecule files is generated. Then, files are converted to the .pdbqt format, using suitable Open Babel classes. After file conversion is done, a loop is initiated to run Vina for every ligand available in the folder and selected macromolecule. Data of each run (10 conformations for each ligand) are saved in a file named ligandname-out.pdbqt (ligandname is the name of the ligand file). After docking rounds are completed, Vina-split is run to split the output files and extract the best iteration. Finally, energy values are read from these files and structures are sorted, by ascending binding energy. Based on user preference, certain number of drug file names and their rank and binding energy are written to a file called Results.txt. This file is shown to the user at the end of the virtual screening process.
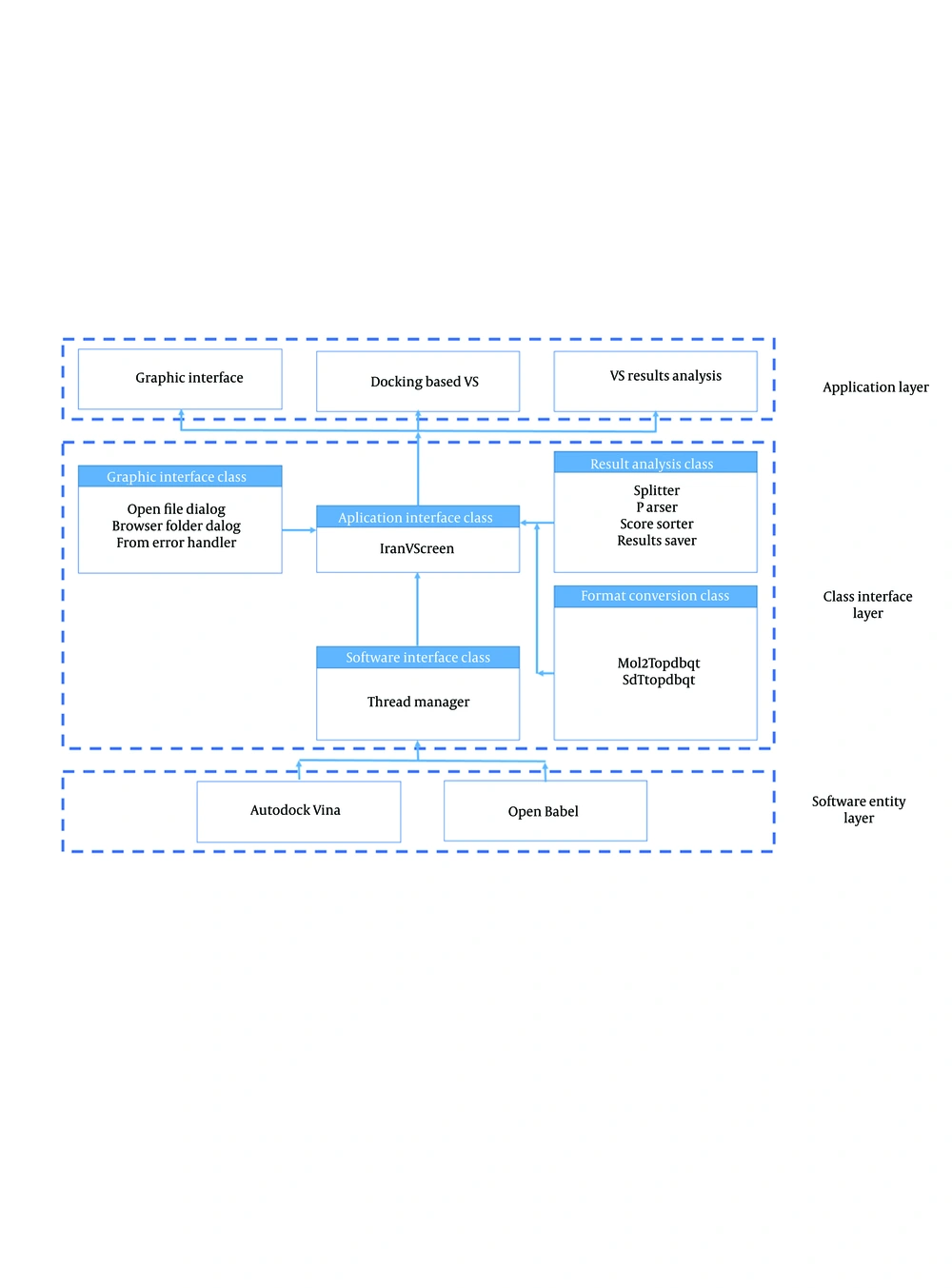
Figure 2 shows that the architecture of the platform consists of three layers: application layer, class interface layer and software entity layer. The class interface layer includes a windows form class, written in Visual Basic, which forms the core of the platform. The integrated software programs are integrated in the software entity layer. The services provided to the user are included in the application layer. Microsoft Visual Studio programming environment provides a uniform software environment for the development of software in different languages, and therefore it was selected to enable authors to easily mix the C++ and Visual Basic codes. The class library in the class interface layer has four components: GUI class, software interface class, results analysis class and format reading class. IranVScreen class is the main class for calling other classes. This class also manages the life cycle of Vina operation thread. The application layer is composed of the GUI and two functional modules. The GUI provides the required data to other modules and displays the results to the user. All data files used by this software are text files and all of the data is written to the work directory, selected by the user.
5. Discussion
In several rare molecule files, an atom record might be used as an alias for a larger group; for example, a molecule might contain an atom named Ph, which is a codename for a phenyl ring. At the time of development, Open Babel could not handle these aliases. A homebrew class was developed to handle such atoms, which was not ideal. Open Babel has added a class named AliasData that processes such files and outputs generic data, since API version 2.2.
The present study describes that IranVScreen provides a very intuitive all-in-one graphic user interface to carry out multiple VS tasks in several mouse clicks, with minimal requirement of skill. The IranVScreen software, localized in Persian language, surpasses the language barrier for novice users.