1. Background
Diabetes mellitus (DM) is a serious public health problem and a devastating condition; its prevalence has increased over the past few decades. It is defined as a group of chronic metabolic dysregulation characterized by hyperglycemia resulting from the lack of insulin production, resistance to insulin action, or even both (1). According to the International Diabetes Federation, the number of diabetic people is expected to exceed 640 million by 2040 (2). Diabetic neuropathy is one of the complications of diabetes (2). Diabetic neuropathy is one of the complications of diabetes. Peripheral neuropathy is defined as peripheral nervous system disorders. Many etiological factors have been involved in the development of peripheral neuropathy, including cancer, drug toxicity, and vitamin deficiencies. The number of patients with DM is growing worldwide; it is one of the most common leading causes of neuropathy, resulting in high morbidity and mortality (3). Diabetic neuropathy includes various neuropathies, including mononeuropathy, polyneuropathy, plexopathy, and radiculopathy (4).
Diabetic neuropathy can produce both painful and non-painful forms. Painful diabetic neuropathy has been estimated to occur in 25% of patients with DM (5, 6). In this regard, the most common form of neuropathic pain arises from type 2 diabetes mellitus (T2DM) (7). In addition, diabetic neuropathy pain (DNP) has been observed in 19% of insulin-dependent patients and 49% of those with non-insulin-dependent DM (8). As diabetes increases, DNP continues to rise with the global diabetes epidemic. Pain is the most common distressing symptom in diabetic neuropathy and mainly affects the lower limbs, including hands and feet. Also, there is a lack of safe and effective sedative drugs to control this chronic painful status. The risk factors causing painful diabetic neuropathy are not as well defined; however, the patient's age, duration of diabetes, nephropathy, peripheral vascular disease, and waist circumference are reported as possible predictors for painful neuropathy progression (9). Research on possible mechanisms involved in diabetic neuropathic pain is very complex because diabetes is a multifactorial disorder. According to the literature, diabetic peripheral neuropathy is associated with hyperglycemia and hyperlipidemia pathology (10). Additionally, diabetic peripheral neuropathy is related to demyelination and degeneration of axons, resulting in nerve dysfunction (7).
It is demonstrated that the soma of the primary afferent neurons that innervate the feet are reported to be present in the lumbar dorsal root ganglia. A dysregulated peripheral nociceptor is involved in promoting pain hypersensitivity in patients with diabetic peripheral neuropathy (11). The proposed causes of dysfunction of nociceptive neurons in the dorsal root ganglia are still being investigated. Studying and modeling complex biological systems to describe various human diseases has attracted much attention in recent years (12). Major biological processes and disease pathogenesis are mediated through physical interactions of proteins; hence, there is a requirement to discover the protein interaction network that forms these processes that leads to understanding human diseases (13). The applications of protein interaction networks allow the identification of genes and proteins related to diseases. Several omics-based investigations were performed on differentially expressed genes (DEGs) in painful diabetic peripheral neuropathy models to identify DEGs contributing to pathological processes and neuropathic pain (14-16). Additionally, network-based analysis can explain the critical genes associated with different diseases (13, 17, 18). Since the full mechanism of painful diabetic neuropathy is not clear, the protein interaction network analysis could be a promising way to manage this problem.
2. Objectives
In the present study, we collected known genes/or proteins related to painful diabetic neuropathy and constructed a network. The important proteins are highlighted as critically involved proteins in pain among patients with diabetic neuropathy.
3. Methods
3.1. Collection of Expression Data Associated with Painful Diabetic Neuropathy
The transcriptomic (genes) and proteomic (proteins) data associated with PDN (painful diabetic neuropathy) were extracted from Web of Sciences, PubMed, Google Scholar, and ScienceDirect using “Painful Diabetic Neuropathy “AND" Differential Protein OR Genes and “Expression Profiling" keywords. The differentially expressed proteins or genes were collected after a literature review and selection of related papers (14, 15, 19, 20).
3.2. Functional Annotation and Pathway Enrichment Analysis
Gene ontology (GO) categories were analyzed to identify the function of genes related to PDN. The GO analysis includes biological processes, molecular function, cellular components, and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis performed using DAVID tools.
3.3. Construction and Analysis of Protein-Protein Interaction Network Related to Painful Diabetic Neuropathy
The Uniporter accession numbers of the collected data were extracted (https://www.uniprot.org). The construction and analysis of the protein-protein interaction (PPI) network were performed using the STRING online web resource and Cytoscape software platform, respectively. The STRING database contains protein interaction data from different sources, including experimental information, computational prediction methods, and public text collections. Cytoscape is an open-source software project for visualization and integrating biomolecular interaction networks with high-throughput expression data (21). The current study analyzed the network characteristics by the Molecular Complex Detection (MCODE) plugin in Cytoscape.
4. Results
4.1. Identification of Differentially Expressed Genes
After the literature review, 4 studies were selected for this analysis. The characteristics of the included studies are shown in Table 1. According to the systematic search, 147 candidate proteins/genes were identified, which include 91 up-regulated and 56 down-regulated proteins/genes (Table 2). For those studies that were performed on rat and mouse models, only the proteins that were common in humans were selected. The threshold of a P-value < 0.05 and FC ≥ 1.5 were considered significantly differentially expressed proteins (Table 2).
| Study Title | Samples | References |
|---|---|---|
| Integrative multi-omic analyses of dorsal root ganglia in diabetic neuropathic pain using proteomics, phosphor-proteomics, and metabolomics | Dorsal root ganglia (DRG) (L4, L5, and S1) from human | Doty et al. 2022 (15) |
| Transcriptomic analysis of human sensory neurons in painful diabetic neuropathy reveals inflammation and neuronal loss. | L4 and L5 ganglia from human | Hall et al. 2022 (19) |
| Proteomics analysis of the spinal dorsal horn in diabetic painful neuropathy rats with electroacupuncture treatment | Spinal dorsal horn sample from a rat model | Yu et al. 2021 (20) |
| Diabetic neuropathic pain induced by streptozotocin alters the expression profile of non‑coding RNAs in the spinal cord of mice as determined by sequencing analysis. | L4‑5 spinal cord tissues from mice model | He et al. 2021 (14) |
| N | UniProt AC. No. | Gene Symbol | Direction of Regulation | N | UniProt AC. No. | Gene Symbol | Direction of Regulation |
|---|---|---|---|---|---|---|---|
| 1 | P02766 | TTR | Up | 75 | P34910 | EVI2B | Up |
| 2 | P62140 | PPP1CB | Up | 76 | P41236 | PPP1R2 | Up |
| 3 | P31213 | SRD5A2 | Up | 77 | E1BB50 | CDK12 | Up |
| 4 | Q969M1 | TOMM40L | Up | 78 | P08670 | VIM | Up |
| 5 | Q96RN1 | SLC26A8 | Up | 79 | Q2TB10 | ZNF800 | Up |
| 6 | Q86Y07 | VRK2 | Up | 80 | P10451 | SPP1 | Up |
| 7 | P35557 | GCK | Up | 81 | Q9C0I1 | MTMR12 | Up |
| 8 | P01042 | KNG1 | Up | 82 | Q13671 | RIN1 | Up |
| 9 | P00738 | HP | Up | 83 | P23588 | EIF4B | Up |
| 10 | Q8IYT4 | KATNAL2 | Up | 84 | O14672 | ADAM10 | Up |
| 11 | P0DOY2 | IGLC2 | Up | 85 | P13639 | EEF2 | Up |
| 12 | Q9H7M9 | VSIR | Up | 86 | Q7Z4Q2 | HEATR3 | Up |
| 13 | O14556 | GAPDHS | Up | 87 | P10915 | HAPLN1 | Up |
| 14 | P02671 | FGA | Up | 88 | Q92752 | TNR | Up |
| 15 | P34982 | OR1D2 | Up | 89 | P48539 | PCP4 | Up |
| 16 | P0DP58 | LYNX1 | Up | 90 | Q7Z4Q2 | HEATR3 | Up |
| 17 | Q99623 | PHB2 | Up | 91 | O43312 | MTSS1 | Up |
| 18 | P02679 | FGG | Up | 92 | Q9UK22 | FBXO2 | Down |
| 19 | P02675 | FGB | Up | 93 | Q9UBY5 | LPAR3 | Down |
| 20 | P35232 | PHB1 | Up | 94 | A5PKU2 | TUSC5 | Down |
| 21 | Q6R327 | RICTOR | Up | 95 | Q5JUK3 | KCNT1 | Down |
| 22 | O94772 | LY6H | Up | 96 | Q9BWW7 | SCRT1 | Down |
| 23 | P21589 | NT5E | Up | 97 | Q8N398 | VWA5B2 | Down |
| 24 | P09669 | COX6C | Up | 98 | Q53GA4 | PHLDA2 | Down |
| 25 | Q9P2U8 | SLC17A6 | Up | 99 | O43711 | TLX3 | Down |
| 26 | P24311 | COX7B | Up | 100 | Q9UHG2 | PCSK1N | Down |
| 27 | O00217 | NDUFS8 | Up | 101 | Q9BQ87 | TBL1Y | Down |
| 28 | P01834 | IGKC | Up | 102 | Q99453 | PHOX2B | Down |
| 29 | P13073 | COX4I1 | Up | 103 | Q9UHR6 | ZNHIT2 | Down |
| 30 | P03915 | MT-ND5 | Up | 104 | Q96A47 | ISL2 | Down |
| 31 | P15954 | COX7C | Up | 105 | Q9H4Q4 | PRDM12 | Down |
| 32 | P17568 | NDUFB7 | Up | 106 | Q02575 | NHLH1 | Down |
| 33 | O43674 | NDUFB5 | Up | 107 | Q9NQ03 | SCRT2 | Down |
| 34 | Q9UI09 | NDUFA12 | Up | 108 | Q9UIU6 | SIX4 | Down |
| 35 | O14548 | COX7A2L | Up | 109 | P12277 | CKB | Down |
| 36 | Q8NA47 | CCDC63 | Up | 110 | P12532 | CKMT1A | Down |
| 37 | Q6ZR08 | DNAH12 | Up | 111 | C9JSQ1 | CKMT1B | Down |
| 38 | Q16864 | ATP6V1F | Up | 112 | Q12809 | KCNH2 | Down |
| 39 | Q96JX3 | SERAC1 | Up | 113 | O43526 | KCNQ2 | Down |
| 40 | Q6ZTW0 | TPGS1 | Up | 114 | Q14654 | KCNJ11 | Down |
| 41 | O75489 | NDUFS3 | Up | 115 | Q92952 | KCNN1 | Down |
| 42 | Q96ID5 | IGSF21 | Up | 116 | Q9NS61 | KCNIP2 | Down |
| 43 | Q9ULK4 | MED23 | Up | 117 | Q9Y2W7 | KCNIP3 | Down |
| 44 | Q9UGC6 | RGS17 | Up | 118 | Q9UHC3 | ASIC3 | Down |
| 45 | P10606 | COX5B | Up | 119 | P30542 | ADORA1 | Down |
| 46 | Q92793 | CREBBP | Up | 120 | P18825 | ADRA2C | Down |
| 47 | Q86UD0 | SAPCD2 | Up | 121 | P41145 | OPRK1 | Down |
| 48 | O43920 | NDUFS5 | Up | 122 | A6NFN3 | RBFOX3 | Down |
| 49 | O95168 | NDUFB4 | Up | 123 | O76070 | SNCG | Down |
| 50 | Q9UKT6 | FBXL21 | Up | 124 | P20472 | PVALB | Down |
| 51 | Q96RD9 | FCRL5 | Up | 125 | Q8N9F0 | NAT8L | Down |
| 52 | A0A0C4DH67 | IGKV1-8 | Up | 126 | Q8N7H5 | PAF1 | Down |
| 53 | A0A0B4J1U7 | IGHV6-1 | Up | 127 | Q96B36 | AKT1S1 | Down |
| 54 | P01599 | IGKV1-17 | Up | 128 | P04035 | HMGCR | Down |
| 55 | A0A0C4DH29 | IGHV1-3 | Up | 129 | Q9C005 | DPY30 | Down |
| 56 | A0A0C4DH69 | IGKV1-9 | Up | 130 | Q9UMX0 | UBQLN1 | Down |
| 57 | P15018 | LIF | Up | 131 | Q0D2I5 | IFFO1 | Down |
| 58 | P01591 | JCHAIN | Up | 132 | Q15819 | UBE2V2 | Down |
| 59 | P01857 | IGHG1 | Up | 133 | Q68D86 | CCDC102B | Down |
| 60 | P06702 | S100A9 | Up | 134 | P07910 | HNRNPC | Down |
| 61 | P05109 | S100A8 | Up | 135 | P36776 | LONP1 | Down |
| 62 | Q9Y5Y7 | LYVE1 | Up | 136 | P02461 | COL3A1 | Down |
| 63 | Q86VB7 | CD163 | Up | 137 | Q13591 | SEMA5A | Down |
| 64 | Q16649 | NFIL3 | Up | 138 | Q9Y5H1 | PCDHGA2 | Down |
| 65 | Q16666 | IFI16 | Up | 139 | P55083 | MFAP4 | Down |
| 66 | P41182 | BCL6 | Up | 140 | Q8N0U8 | VKORC1L1 | Down |
| 67 | Q6W2J9 | BCOR | Up | 141 | Q8WUX1 | SLC38A5 | Down |
| 68 | Q08881 | ITK | Up | 142 | P22692 | IGFBP4 | Down |
| 69 | P41279 | MAP3K8 | Up | 143 | P02462 | COL4A1 | Down |
| 70 | P24941 | CDK2 | Up | 144 | Q9BX70 | BTBD2 | Down |
| 71 | O60462 | NRP2 | Up | 145 | P15880 | RPS2 | Down |
| 72 | P23588 | EIF4B | Up | 146 | Q7Z6Z7 | HUWE1 | Down |
| 73 | O14672 | ADAM10 | Up | 147 | P02649 | APOE | Down |
| 74 | P13639 | EEF2 | Up |
a P-value < 0.05 and fold change > 1.5.
4.2. Functional and Pathway Enrichment Analyses
According to Table 3, in the biological processes-associated category, the DRGs significantly enriched mitochondrial respiratory chain Complex I assembly (GO: 0032981, P-value = 2.1E-4), mitochondrial ATP synthesis coupled proton transport (GO: 0042776, P-value = 2.1E-4), aerobic respiration (GO: 0009060, P-value = 2.1E-4), etc. The molecular function annotation results of DEGs included NADH dehydrogenase (ubiquinone) activity (GO: 0008137, P-value = 1.2E-2), antigen binding (GO: 0003823, P-value = 1.2E-2), receptor binding (GO: 0005102, P-value = 5.9e-01), cytochrome-c oxidase activity (GO: 0004129, P-value = 2.8E-2), etc. In addition, the cellular component annotation indicated that the mitochondrial inner membrane (GO: 0005743, P-value = 3.0E-6), mitochondrial respiratory chain Complex I (GO: 0005747, P-value = 3.6E-6), and blood microparticle (GO: 0072562, P-value = 3.6E-6) were major enriched categories in DEGs.
| Term | Count | Benjamini-Corrected P-Value | Fold Enrichment |
|---|---|---|---|
| GOTERM: Biological process | |||
| GO:0032981~mitochondrial respiratory chain complex I assembly | 8 | 2.1E-4 | 17.76 |
| GO:0042776~ mitochondrial ATP synthesis coupled proton transport | 8 | 2.1E-4 | 17.22 |
| GO:0009060~ aerobic respiration | 8 | 2.1E-4 | 15.99 |
| GO:0006123~ mitochondrial electron transport, cytochrome c to oxygen | 6 | 2.1E-4 | 33.57 |
| GO:0006120~ mitochondrial electron transport, NADH to ubiquinone | 7 | 2.1E-4 | 20.83 |
| GO:0045907~ positive regulation of vasoconstriction | 5 | 2.7E-2 | 18.40 |
| GO:0045333~ cellular respiration | 5 | 3.5E-2 | 16.65 |
| GO:0071805~ potassium ion transmembrane transport | 7 | 4.6E-2 | 7.53 |
| GOTERM: Molecular function | |||
| GO:0008137~ NADH dehydrogenase (ubiquinone) activity | 8 | 6.0E-6 | 26.19 |
| GO:0003823~ antigen binding | 8 | 1.2E-2 | 7.76 |
| GO:0005102~ receptor binding | 12 | 1.2E-2 | 4.25 |
| GO:0004129~ cytochrome-c oxidase activity | 4 | 2.8E-2 | 28.16 |
| GO:0034987~ immunoglobulin receptor binding | 6 | 3.7E-2 | 8.80 |
| GO:0004111~ creatine kinase activity | 3 | 3.8E-2 | 70.40 |
| GOTERM: Cellular component | |||
| GO:0005743~ mitochondrial inner membrane | 18 | 3.0E-6 | 3.4 |
| GO:0005747~ mitochondrial respiratory chain complex I | 8 | 3.6E-6 | 2.9 |
| GO:0072562~ blood microparticle | 11 | 3.6E-6 | 2.4 |
| GO:0005751~ mitochondrial respiratory chain Complex IV | 6 | 4.2E-5 | 1.6 |
| GO:0045202~ synapse | 14 | 9.2E-4 | 1.4 |
| GO:0005739~ mitochondrion | 25 | 9.2E-4 | 1.0 |
| GO:0031966~ mitochondrial membrane | 8 | 3.2E-3 | 1.1 |
| GO:0070062~ extracellular exosome | 30 | 1.1E-2 | 1.1 |
| GO:0043025~ neuronal cell body | 11 | 2.1E-2 | 0.8 |
The KEGG pathway analysis showed that oxidative phosphorylation (P-value = 2.7E-9), diabetic cardiomyopathy (P-value = 3.5E-8), and non-alcoholic fatty liver disease (P-value = 9.1E-7) were the mainly enriched pathways associated with DRGs in painful diabetic neuropathy, as presented in Table 4.
| KEGG Pathway | Count | Benjamini-Corrected P-Value | Fold Enrichment |
|---|---|---|---|
| Oxidative phosphorylation | 15 | 2.7E-9 | 11.71 |
| Diabetic cardiomyopathy | 16 | 3.5E-8 | 8.24 |
| Non-alcoholic fatty liver disease | 13 | 9.1E-7 | 8.77 |
| Amyotrophic lateral sclerosis | 15 | 9.7E-7 | 6.76 |
| Thermogenesis | 16 | 3.3E-6 | 5.47 |
| Huntington disease | 14 | 3.3E-6 | 6.57 |
| Chemical carcinogenesis - reactive oxygen species | 17 | 4.3E-6 | 4.88 |
| Parkinson's disease | 10 | 2.2E-5 | 5.36 |
| Prion disease | 12 | 3.4E-5 | 4.36 |
| Alzheimer's disease | 10 | 1.1E-5 | 3.51 |
| Pathways of neurodegeneration - multiple diseases | 17 | 4.1E-4 | 6.36 |
| Metabolic pathways | 25 | 6.8E-2 | 1.69 |
Abbreviation: KEEG, Kyoto Encyclopedia of Genes and Genomes.
a The count threshold was ≥ 10.
4.3. Identification of Hub Genes via PPI Network Analysis
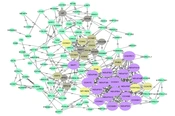
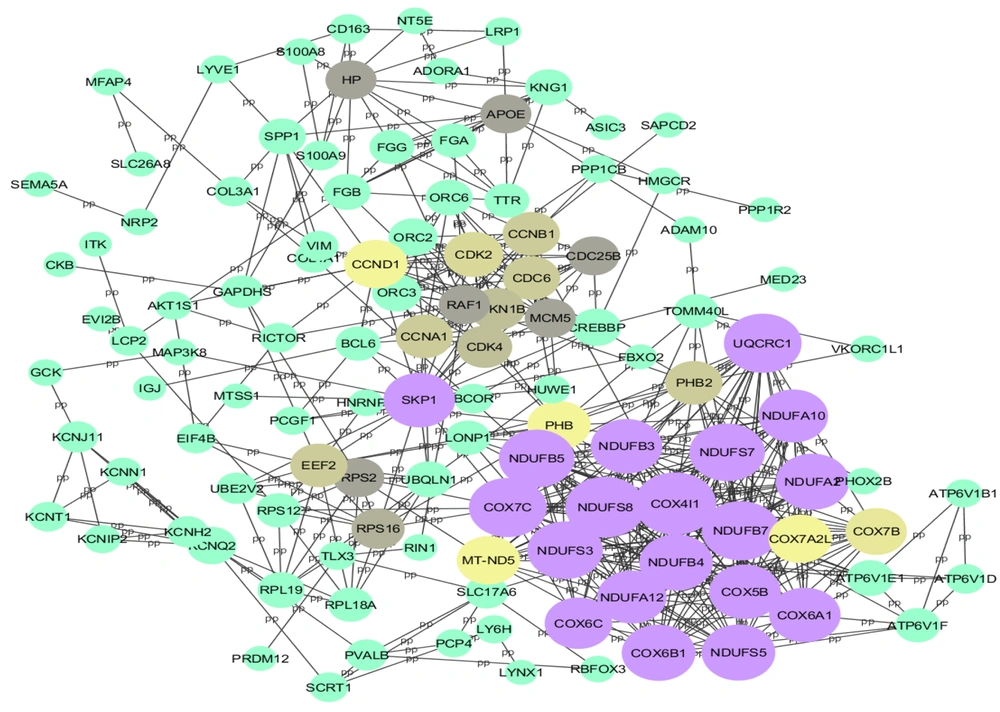
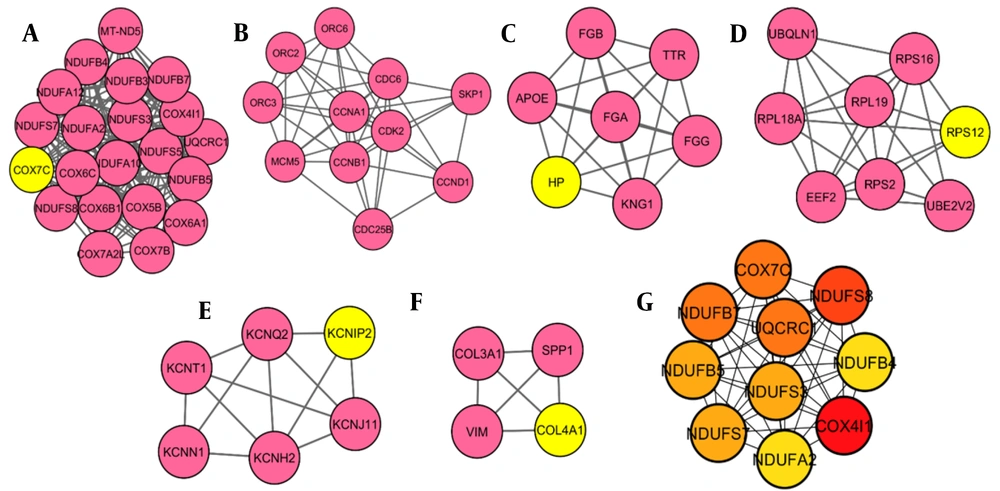
After removing disconnected nodes, the STRING database yielded a PPI network with 117 nodes. The network was then analyzed in Cytoscape software, shown in Figure 1. Hub proteins were selected according to CytoHubba, and 6 distinct clusters were extracted from the network using the MCODE plugin (Figure 2A - F). The results showed that the most important hub proteins in the network included COX4I1, NDUFS8, UQCRC1, COX7C, and some other NADH dehydrogenases, including NDUFB7, NDUFS7, NDUFS3, NDUFB5, NDUFA2, and NDUFB4. The results of the functional enrichment analysis of the clusters showed that the most important pathways that the clusters (clusters 1 - 5) were involved in oxidative phosphorylation, cell cycle, complement, and coagulation cascades, ribosome, and GnRH secretion (Table 5), while no significant KEGG pathways detected for Cluster 6.
| KEGG Pathway | Count | Benjamini-Corrected P-Value |
|---|---|---|
| Oxidative phosphorylation (cluster 1) | 21 | 6.5E-36 |
| Cell cycle (cluster 2) | 11 | 3.3E-17 |
| Complement and coagulation cascades (cluster 3) | 4 | 1.7E-4 |
| Ribosome (cluster 4) | 5 | 3.2E-5 |
| GnRH secretion (cluster 5) | 2 | 4.2E-2 |
Abbreviation: KEEG, Kyoto Encyclopedia of Genes and Genomes.
5. Discussion
Since the proteins interact with each other in the cellular pathways, many disorders result from the deregulation of proteins. The protein interaction network-based analysis is beneficial for systematically studying complex and multifactorial diseases such as cancer and DM (22). Protein-protein interactions contribute to all vital biological activities in living organisms. Identifying protein interactions in the cells is essential to reveal the function and cellular and molecular mechanisms in cells. Commonly, PPI can provide a valuable overview for a great comprehension of the functional organization of the proteome. This modern approach is now used as an efficient method to identify potential drug, therapeutic, diagnostic, and prognostic targets in various diseases (17, 23). An important advantage of network analysis is the identification of hub nodes in the protein interaction network. In the present study, the protein interaction network associated with painful diabetic neuropathy was constructed and evaluated. We extracted 147 proteins and genes with differential expression from literature and predicted the main proteins as potential biomarkers related to peripheral PDN. The top 10 nodes (hub proteins), which mostly interact with the other nodes, are represented in the result section, include COX4I1, NDUFS8, UQCRC1, COX7C, NDUFB7, NDUFS7, NDUFS3, NDUFB5, NDUFA2, and NDUFB4. These proteins were identified as the essential proteins that play critical roles in pathophysiology and cellular pathways related to pain in diabetic neuropathy.
In this study, COX4I1 and COX7C were identified as hub proteins with the highest degree. Cytochrome c oxidase (COX) is an indispensable part of mitochondrial machinery needed for ATP production in mammalian cells. In addition to 3 mitochondria-encoded subunits necessary for COX catalytic function, 11 nuclear-encoded subunits build up the COX enzyme and regulate COX enzyme activity. Cytochrome c oxidase is regulated via tissue-, development- or environment-controlled expression of subunit isoforms. The COX4 subunit is thought to optimize respiratory chain function based on the oxygen-controlled expression of its isoforms COX4I1 and COX4I2 (24). Studies show low COX4I1 links mitochondrial dysfunction to obesity and T2DM in humans and mice (25). Dysregulation of the COX complex is related to mitochondrial oxidative stress (26). In addition, the oxidative stress condition in mitochondria is associated with obesity, metabolic syndrome, and T2DM (27). COX4I1 is suggested to be the most important regulatory subunit of COX (28). Van der Schueren et al. (25) conducted a study to investigate the association of mitochondrial oxidative stress with obesity, metabolic syndrome, and T2DM and evaluate COX4I1 in peripheral blood monocytes as well as a potential biomarker for harmful metabolic development in obesity patients. They reported that COX4I1 depression is associated with insulin resistance and T2DM in obesity. Moreover, it is perhaps a helpful diagnostic biomarker in peripheral blood monocytes (25). Another study reported that low cytochrome oxidase1 links mitochondrial dysfunction to atherosclerosis (29). Recently, a study analyzed the proteomics of the spinal dorsal horn in diabetic painful neuropathy rats, and their results indicated that COX (COX, Complex IV) factors, including COX4I1, COX5B, COX6C2, COX7B, and COX7C, were significantly up-regulated in spinal dorsal horn during PDN (20). Besides, the COX7C is not only a hub but also recognized as a seed node, which shows its importance in the pathogenesis of neuropathy as well as a potential drug target.
In our study, NDUFS8 is detected as another hub protein. The NDUFS8 protein is a subunit of NADH dehydrogenase (ubiquinone), also called Complex I, that is located in the inner membrane of mitochondria. Mutations in NDUFS8 have been associated with clinical features, including ptosis, external ophthalmoplegia, proximal myopathy, cardiomyopathy, pigmentary retinopathy, encephalopathy, and neurodegenerative disorders. Type 1 diabetes mellitus (T1DM) is an endocrine disorder characterized by destroying pancreatic β cells. This is attributed to the development of chronic diabetic complications: neurovascular and macrovascular. The development of complications is associated with various risk factors, mainly insulin resistance (30, 31) and hyperglycemia. Flotynska et al. conducted a study to evaluate NDUFS8 serum concentration as a Complex I marker and the relationship with insulin resistance in T1DM. It has been found that a higher serum concentration of NDUFS8 protein is associated with higher insulin sensitivity among adult patients with T1DM (32). In addition, the NDUFS8 gene was expressed at a high level in the skeletal muscle tissue of T2DM patients, which might indicate that increased expression of NDUFS8 can affect the glucose metabolism in the skeletal muscle tissue, causing insulin resistance and then diabetes development. Furthermore, based on bioinformatics analysis, NDUFS8 is a potential therapeutic target (33).
Another hub protein in our analysis is UQCRC1, characterized as a subunit of Complex III in the mitochondrial respiratory chain. The functional effect of UQCRC1 mutations was investigated in several study models to assess their potential pathogenicity in the disease process. In this regard, it is demonstrated that the mitochondrial UQCRC1 mutations cause autosomal dominant Parkinsonism with polyneuropathy (34). Although the important role of mitochondria in the development of diabetes and its complications, especially neuropathy, is evident, there have been fewer studies on the physiology of this organelle in diabetic neuropathy than in other complications such as cardiomyopathy. According to this, in one study of alterations in mitochondrial physiology, the mRNA level of UQCRC1 decreased in the Diabetic Akita mouse model (35).
Interestingly, some other NADH dehydrogenases, including NDUFB7, NDUFS7, NDUFS3, NDUFB5, NDUFA2, and NDUFB4, are detected as hub proteins in our network analysis. Diabetic neuropathy is a main complication of DM that causes significant morbidity among patients with diabetes. Meanwhile, mitochondrial dysfunction and oxidative stress have been suggested as important mediators of neurodegeneration in diabetes (36). It is suggested that high glucose in tissues triggers extreme electron donation to the electron transport chain and an elevated supply of NADH in the mitochondria, leading to increased reactive oxygen species and degeneration of target tissues (37). Considering the importance of mitochondria and their cellular processes in diabetes and its complications, we found that oxidative phosphorylation is a significant pathway involved in painful diabetic neuropathy through KEGG pathway analysis. In agreement with the results obtained from our study, several studies also demonstrated that mitochondrial dysfunction occurred in neuropathy (38, 39). In the current study, 6 clusters and 5 seed nodes were also determined through protein network analysis, including COX7C, HP, RPS12, KCNIP2, and CoL4A1, which can serve as candidate biomarkers for painful diabetic neuropathy. However, further investigations are needed to evaluate these proteins in detail.
5.1. Conclusions
Our study intended to detect the main proteins and genes involved in painful diabetic neuropathy progression and identify potential biomarkers using comprehensive bioinformatics analyses. Collectively, 147 differentially regulated (91 up- and 56 down-regulated) proteins/genes were identified in painful diabetic neuropathy. Our research has provided new points into PDN pathogenesis by analyzing the DEG proteins/genes and their interactions with each other and presenting their hub proteins, pathways, and functional annotation. These proteins, including COX7C, HP, RPS12, KCNIP2, and CoL4A1, can be candidate biomarkers and targets for PDN management and potential treatment.