1. Background
Mucormycosis is an invasive and rapidly progressing fungal infection with significant mortality that occurs in immunocompromised patients, and its clinical characteristics and treatment methods are incomplete. Mucormycosis is a term that refers to infections caused by mucoral fungi, such as Rhizopus and Rhizomucor.
After Aspergillus species, mucormycetes are the second most common cause of mycosis in hematological patients (1, 2). Therefore, finding a fast and accurate method to identify mucor species is very important, especially in its clinical samples (3, 4). For this reason, the use of new, fast, and sensitive methods, including molecular methods of deoxyribonucleic acid (DNA) detection in various samples, such as serum, BAL, and tissue, is currently being investigated (5, 6).
Mucormycosis has various clinical manifestations, the most common of which is the classical form of rhinocerebral-orbital. Successful treatment of mucormycosis is performed with a timely diagnosis. For this reason, the accurate diagnosis of the underlying factors seems to be very necessary to determine the appropriate clinical protocol and the prognosis of the disease. Meanwhile, molecular methods give us more accurate results due to high sensitivity and specificity.
Molecular tracking of mucormycosis is conducted based on different methods, such as polymerase chain reaction (PCR) and real-time PCR. Real-time PCR is one of the best molecular detection techniques, primarily due to the reduction of time, and is suitable for large-scale work (7-9). Therefore, the PCR method is used in the diagnosis and follow-up of the treatment of many diseases, such as cancer (10-14).
2. Objectives
In this study, real-time PCR was used to identify mucor and some of its species in clinical samples.
3. Methods
This was a cross-sectional-descriptive study in 1402. After obtaining permission from the Ethics Committee (IR.SBMU.NRITLD.REC.1402.219), 80 tissue samples were collected from referring patients for fungus diagnosis with the opinion of a specialist doctor.
The inclusion criteria for the study were pathology (smear or histopathology) and clinical signs consistent with rhino-orbital-cerebral mucormycosis. The exclusion criteria were the failure to complete the hospital file accurately and the patients who, for any reason, were not given diagnostic and therapeutic measures and left the hospital.
In this study, real-time PCR and PCR methods were employed as molecular techniques for diagnosis. For this purpose, DNA extraction must be performed first. There are different methods for DNA extraction. Commercial DNA extraction kits were utilized to obtain better results and save time. Deoxyribonucleic acid extraction steps were performed using a GeNet Bio kit, and DNA quality and purity were evaluated by NanoDrop.
Before performing real-time RT-PCR and PCR, a specific primer was prepared for mucoral group fungi, and the specifications of the primer are shown in Table 1. To determine efficiency, a real-time RT-PCR reaction was performed with different dilutions of the primer, and the slope of the standard curve was obtained, indicating the primer's efficiency.
| Parameters | Universal-Pan | Mucor |
|---|---|---|
| Forward primer | GCAGYAGATCTCTTAGATCTGA | CTACGACAGTCACATTGGTG |
| Length | 22 | 20 |
| Reverse primer | AACGTTCGTTGACTCAATGG | ATCGGATCTAGAGATCTAGTC |
| Length | 20 | 21 |
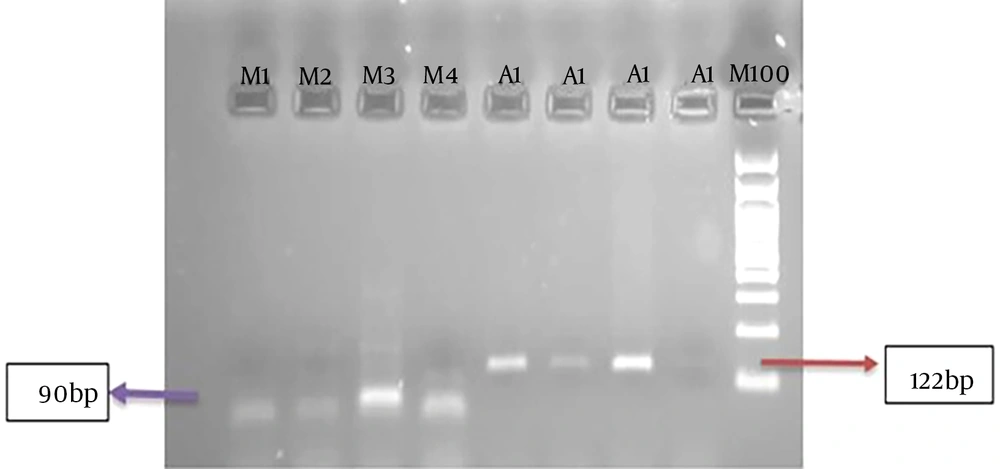
| product length | 122 | 90 |
| Annealing | 62 | 64 |
Using databases and the powerful Blast tool confirmed the binding site of the primer pair to the relevant gene. Primers were made by Gene Fanavaran company (Iran). The samples were analyzed by real-time PCR using specific primers to identify mucor (Table 1).
Real-time PCR was conducted in a final volume of 20 mL with the content of 12 mL Ampliqon master mix, 0.3 mL of each of F and R primers, 2 µL DNA, and distilled water. By employing a rotor-gene cycler, the test was carried out in 40 cycles.
Temperature and time conditions included an initial denaturation at 95°C for 5 minutes, denaturation at 95°C for 15 seconds, primers connection at 56°C for 60 seconds, and amplification at 72°C for 25 seconds (for 40 cycles). The final amplification was performed at 72°C for 5 minutes. Sequencing was conducted for several samples for final approval.
3.1. Statistical Analysis
The sample size was estimated according to the favorable ratio of the tests and considering 5% type 1 error and 20% type 2 error. The results were analyzed using SPSS software (version 22), and the mean and standard deviation were calculated. A paired t-test was utilized for data analysis. A P-value ≤ 0.05 was regarded as statistically significant.
4. Results
This study evaluated 80 tissue samples that were suspected of fungi in the pathology investigations. The average age of the patients was 56.8 ± 8.6 years. All the samples were cultured on Sabouraud dextrose agar (SDA). A total of 76 samples were culture-positive, and 14 of them were reported to be mucor-positive by examining microscopic smears and specific cultures.
Deoxyribonucleic acid extraction was performed from tissue samples and cultured colonies. Moreover, the molecular test was performed using the PCR method with the pan-fungi-specific primer. The PCR method of the DNA obtained from the colonies confirmed 74 cases of fungi. In addition, the PCR test of the DNA obtained from the tissues showed that 75 patients were positive for fungi. Then, using a specific primer for mucor, a PCR test was performed, and 12 mucor-positive cases were reported in the DNA obtained from the colonies and in the DNA obtained from the tissues.
In the next step, a molecular test was performed using the pan-fungi-specific primer by real-time PCR method. The real-time PCR method of DNA obtained from colonies confirmed 76 cases of fungi. Furthermore, with real-time PCR tests of DNA obtained from tissues, 77 cases were positive for fungi.
Then, a real-time PCR test was performed using a specific primer for mucor, and 16 mucor-positive cases were reported in the DNA obtained from the colonies. Additionally, 15 mucor-positive cases were reported in the DNA obtained from the tissues.
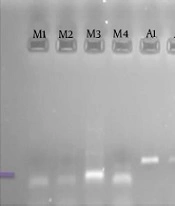
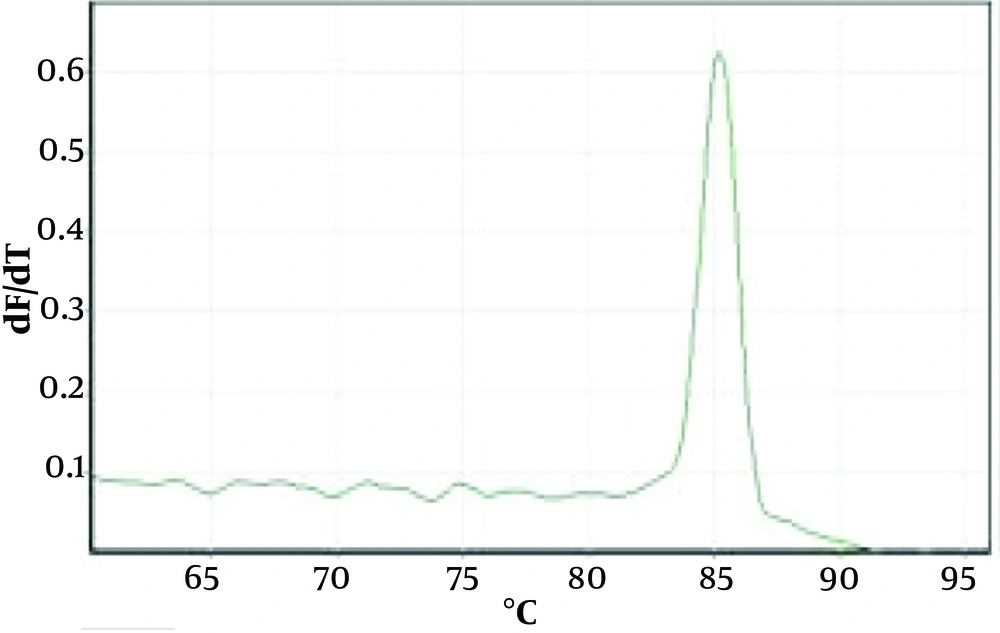
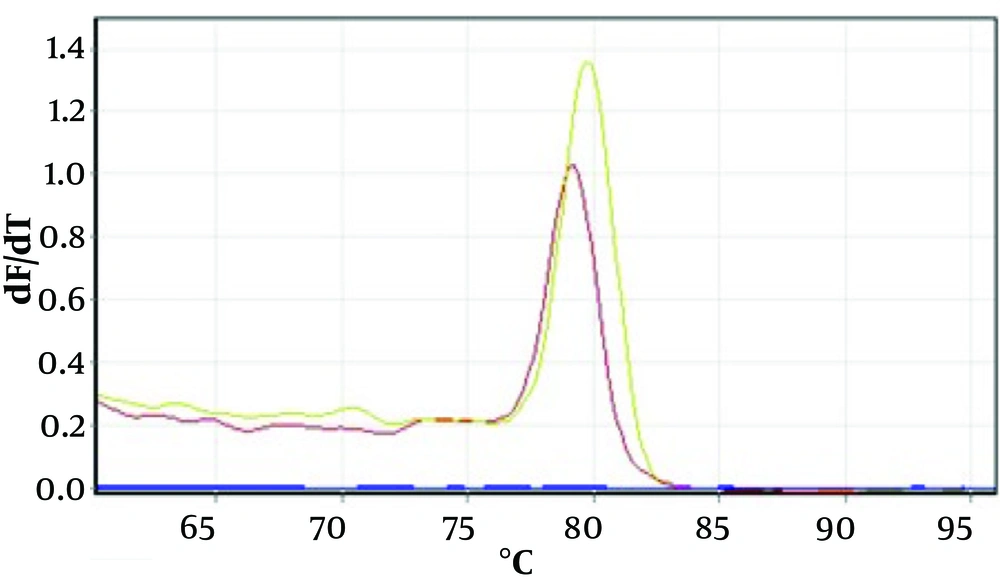
Figure 1 shows an example of the PCR method for agarose gel. Figures 2 and 3 show the melting curves of the real-time PCR method for pan-fungi and mucor.
In the next step, the products of the positive samples from real-time PCR were sent to Gene Fanavaran for sequencing, and the results were compared to the results of real-time PCR tests. The sequence results confirmed the number of 12 cases of real-time PCR results in the case of mucor.
5. Discussion
In recent years, the problems of increasing treatment-resistant fungal organisms on the one hand and the lack of definitive diagnostic methods and tools on the other hand have caused concern about the timely diagnosis and progress of invasive fungal diseases (6). In recent decades, there have been various molecular methods to identify fungi and other diseases (15).
The real-time PCR method, which is used in clinical samples, is one of the best methods to identify and diagnose mucor. In the present study, 80 samples were examined, and PCR confirmed 74 and 75 cases of fungi from DNA obtained from colonies and tissues, respectively. By using specific primers and PCR tests for mucor, 12 cases were mucor and were reported as positive.
In addition, 76 and 77 cases of fungi were confirmed in the real-time PCR method from the DNA obtained from the colonies and tissues, respectively. By the use of specific primers and the real-time PCR test for mucor, 16 cases of mucor and 15 positive cases were reported from the tissues. These differences might be due to the different growth conditions of some fungi in the culture medium, the use of other substances in pathological staining, the use of other DNA extraction kits with different sensitivity, or different mixes used in the PCR process and/or real-time PCR (16).
In a study by Sani et al., the clinical symptoms of the ear, nose, and throat nearly 70% included most of the clinical signs, which is in line with the results of the present study, and the patients had similar clinical signs and were positive for mucor in laboratory tests (17).
It seems that the cause of mucormycosis is different in different cultures, such as mucor spp. and Phizopus spp. (18). Therefore, further research on different molecular diagnostic methods for different causes of mucormycosis and rare pathogens is needed to be able to identify both mucormycosis agents and their subspecies (19).
Millon et al.'s study showed that micromycetes DNA could be identified in at least 81% of patients with possible mucormycosis (20). In a recent study, 80/77 (96%) samples were diagnosed with mucor.
In any case, considering the results and efficiency of molecular methods in a short time in timely diagnosis for microbial and fungal infections, these tests are recommended for medical centers because these molecular methods have good sensitivity and specificity compared to other methods. However, further studies with larger samples and other specific primers are suggested to obtain results and accurately identify mucor factors and their subspecies.