1. Background
Tuberculosis (TB) is a medical and public health issue that needs prevention and treatment despite the increasing rate of multidrug-resistant (MDR) strains that exacerbate the public health problems in the world (1, 2). Many of the newly found medications prescribed for TB have been commonly used for 50 years or even more. Due to new mechanisms of resistance, new genes have emerged and worsened the situation of TB treatment. Thus, new strains are more resistant to medications prescribed for the treatment of TB. Recent reports on antimicrobial resistances worldwide indicate that twenty percent of new TB patients are at least resistant to one drug (TB-DR), 5% were resistant to multi-drug (TB-MDR), and 11.3% showed resistance to streptomycin (S+) (3, 4). The Streptomycin drug, a member of the aminoglycoside that was found in 1943, introduced as the first medication with proven effects on TB, and used in the treatment of pulmonary TB for more than 7 decades. It worked by inhibiting the growth of the bacteria responsible for the infection. However, with the widespread use of the drug, bacteria quickly developed resistance, rendering it ineffective. Drug-resistant TB is caused by bacteria that are resistant to the effects of one or more of the anti-tuberculosis drugs, making treatment challenging and lengthy. In some cases, patients may require hospitalization and the use of more potent medications with severe side effects (5). The mechanism of Streptomycin is acting on the ribosome, inhibiting mRNA translation and thereby disrupting protein production by the organism at extracellular medium, binds to the 12S subunits and 16S rRNA. Resistance to streptomycin among tuberculosis is mainly related to rrs and rpsL genes. The rpsL gene is responsible for the 12S subunit protein, with substitutions in codons of 88 (A/G/C, Lys→Gln, Arg, Thr) and 43 (A/G, Lys→Arg, Thr) predominant. This rrs gene is responsible for 16S rRNA, and its most common mutation sites occur in the loop of 530 and region 912 (6-8). Also, kanamycin and amikacin are aminoglycosides that bind to bacterial ribosomes and inhibit protein synthesis. Aminoglycosides connect to the subunits and make it wrong to read genetic codes and thus increase proteins that have incorrect folding in the cell and, ultimately, cause the cell's death. The nucleotide changes in the rrs gene in the codons of 1400 (A to G), 1401 (C to A), and 1483 (G to T) are associated with resistance to Aminoglycosides. In particular, resistance to high levels of amikacin and kanamycin is associated with mutations in codon 1400 (9, 10). In spite of vigorous advances in the detection of rpsL and rrs mutations in S+ tuberculosis strains, there is insufficient information about these genes, but important geographical diversity identified for the mentioned mutations (3, 11). Regarding the geographical situation for Iran and its neighbor countries with a high prevalence of tuberculosis and due to the migration of refugees and other populations, the current investigation conducted to identify the frequency of spot rrs and rpsL mutations in aminoglycoside resistant strains (kanamycin, amikacin, streptomycin). To obtain data, M.tuberculosis MDR samples taken from clinics belong the western of Iran in 2018 - 2019, to determine the frequency of resistance, the best treatment method, and to prevent the spread of resistance.
2. Methods
2.1. Bacterial Strains and Sample Collection
This descriptive, cross-sectional study was performed on 40 samples of Mycobacterium tuberculosis from January 2018 to June 2019 on suspected tuberculosis patients referred to the west of Iran TB reference laboratory in Kermanshah. In this study, 17 proven drug-resistant samples for aminoglycosides, and 23 susceptible samples were studied. Sputum samples from suspected tuberculosis patients were collected, and if the smear was positive or if the patients exhibited at least one of the signs of prolonged coughing, excessive sputum, hemoptysis, chest pain, decreased appetite, weight loss, fever, and shivering, they were enrolled in the study. For the homogenization and decontamination of samples, sputum was exposed to 4% sodium hydroxide for 30 minutes and then placed under the laminar hood for 15 minutes. After proper exposure, the specimens were centrifuged at 3000 rpm for 15 minutes. Once the supernatant was removed, the precipitate was neutralized with 1 molar neutral chloride acid, and then 3 to 4 drops of the neutralized sediment were inoculated. After cultivation of the specimens, the cultured media were incubated at 37 ºC. After three days, the contaminated tubes were discarded, and samples were taken again. The remaining tubes were reported as having fast growth if Mycobacteria grew during 7 - 10 days. After one week, colonies of colorless to creamy were observed in the culture media (12).
2.2. DNA Extraction
Manual methods are time-consuming, and the accuracy of these methods is controversial. For our investigation, we chose the Extraction Kit. All DNA extraction steps, according to the G-spin Total kit (Cat.NO.17045 South Korea's intron corporation), were set up in the Center for Pulmonary Diseases (Tuberculosis), and highly sterile and safe conditions were considered (9).
2.3. Polymerase Chain Reaction Test (PCR)
The polymerase chain reaction (PCR) was implemented via the AID (GeneXpert/RIF Kit U.S.A) kit, and the other materials, including MgCl2, 10x Buffer, and Taq polymerase, were supplied by Arian Genesis Manufacturing Company. The reactions were conducted in a final volume of 25μL (prepared according to the manufacturer's protocol), including mastermix and 5 μl of extracted DNA. After these steps, thermocycler amplification was performed at a temperature of 95°C for 5 minutes, followed by 14 cycles (30 seconds at 95°C, 2 minutes at 60°C), as well as 26 cycles of 10 seconds at 95°C, 30 seconds at 55°C, 30 seconds at 72°C, and finally at 72°C for 8 minutes.
2.4. Hybridization
It is also possible to perform these steps manually using protocols; however, in order to save time, ensure safety, and achieve reliable data and accurate results, this step was conducted using the molecular Kit Mycobacterium-resistance RDB 2184, manufactured by the German AID Company.
2.5. Analysis
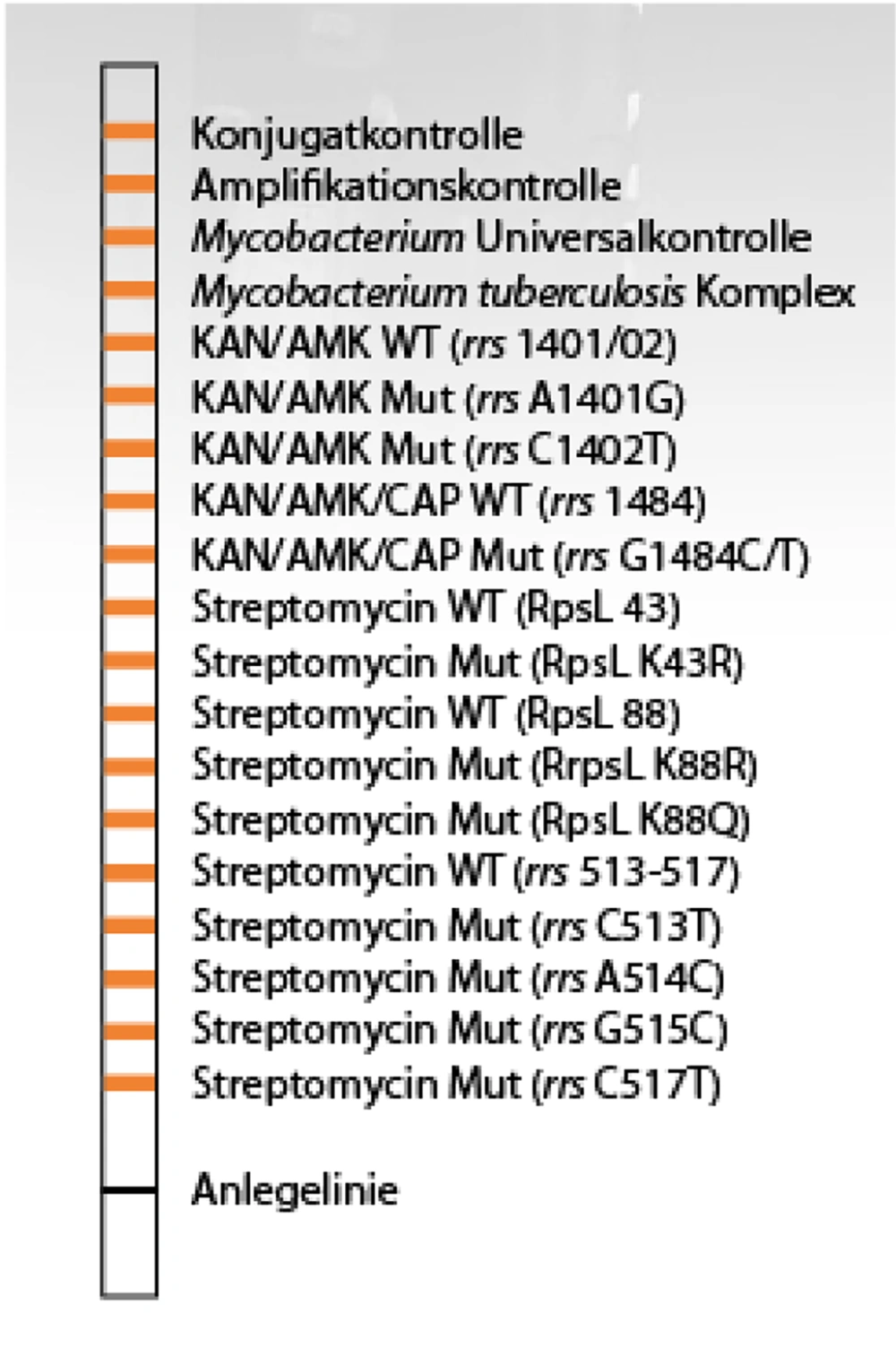
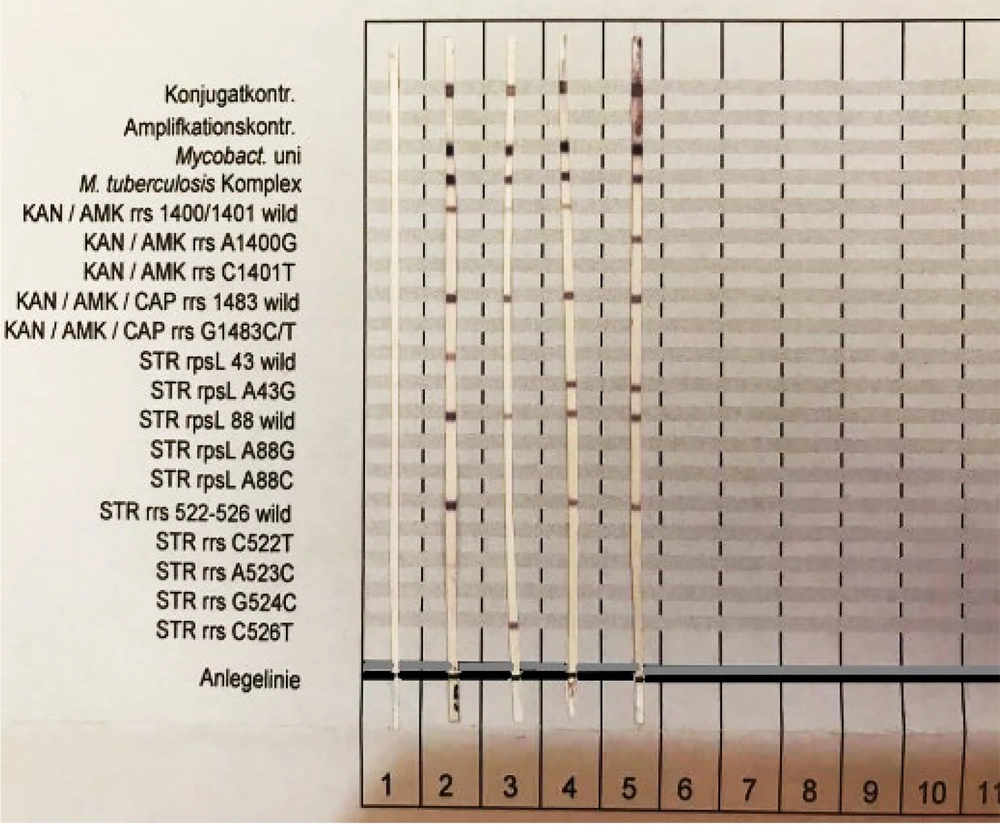
To analyze the status of the reactions created on the strips, the exact position as indicated on the evaluation sheet was determined. The image of a bar and the positions of the mutated genes and wild type are depicted in Figure 1.
3. Results
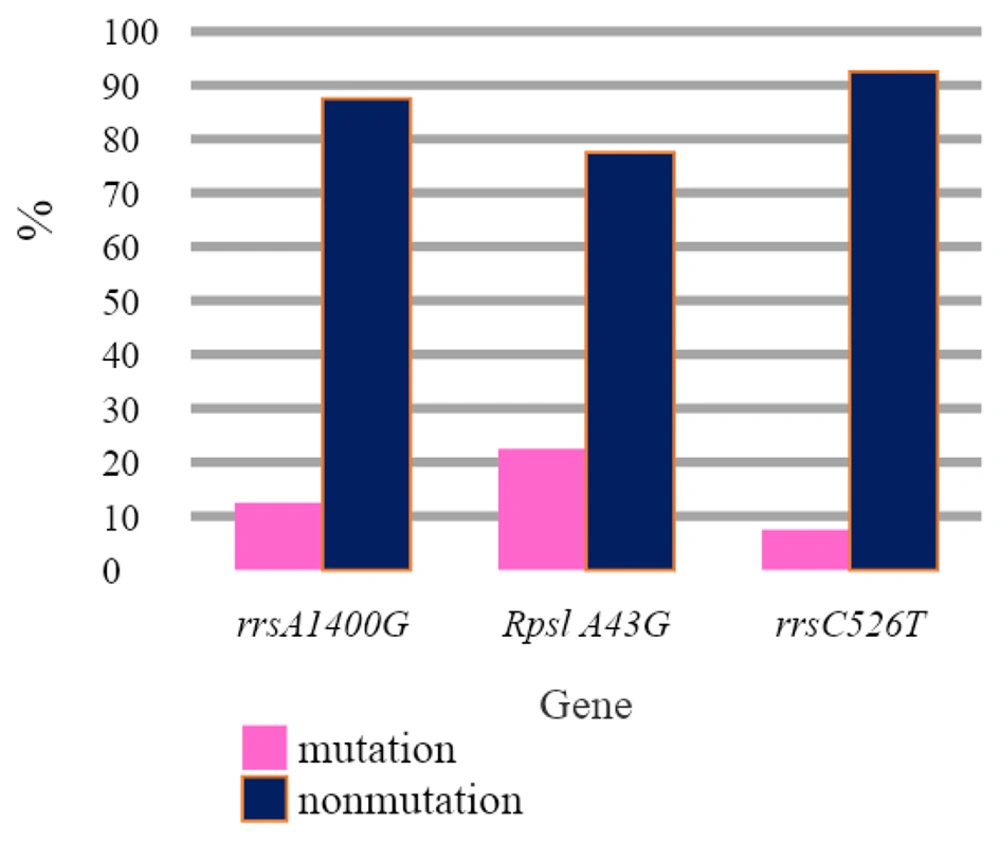
Examination of available strains revealed that five samples, with a mutation frequency of 12.5%, were found in the codon of the 1400 rrs gene, while 87.5% of the samples did not show any mutations in this gene and were susceptible to amikacin and kanamycin. Other samples, with a mutation frequency of 22.5% in the rpsL A43G gene, were identified as resistant to streptomycin antibiotics, but 31 cases (77.5%) did not show mutations in the rpsL gene at codon 43. Three samples, with 7.5% mutations in the rrsC526T gene, were resistant to streptomycin (Figure 2).
3.1. Results of Strains with Similar Mutations
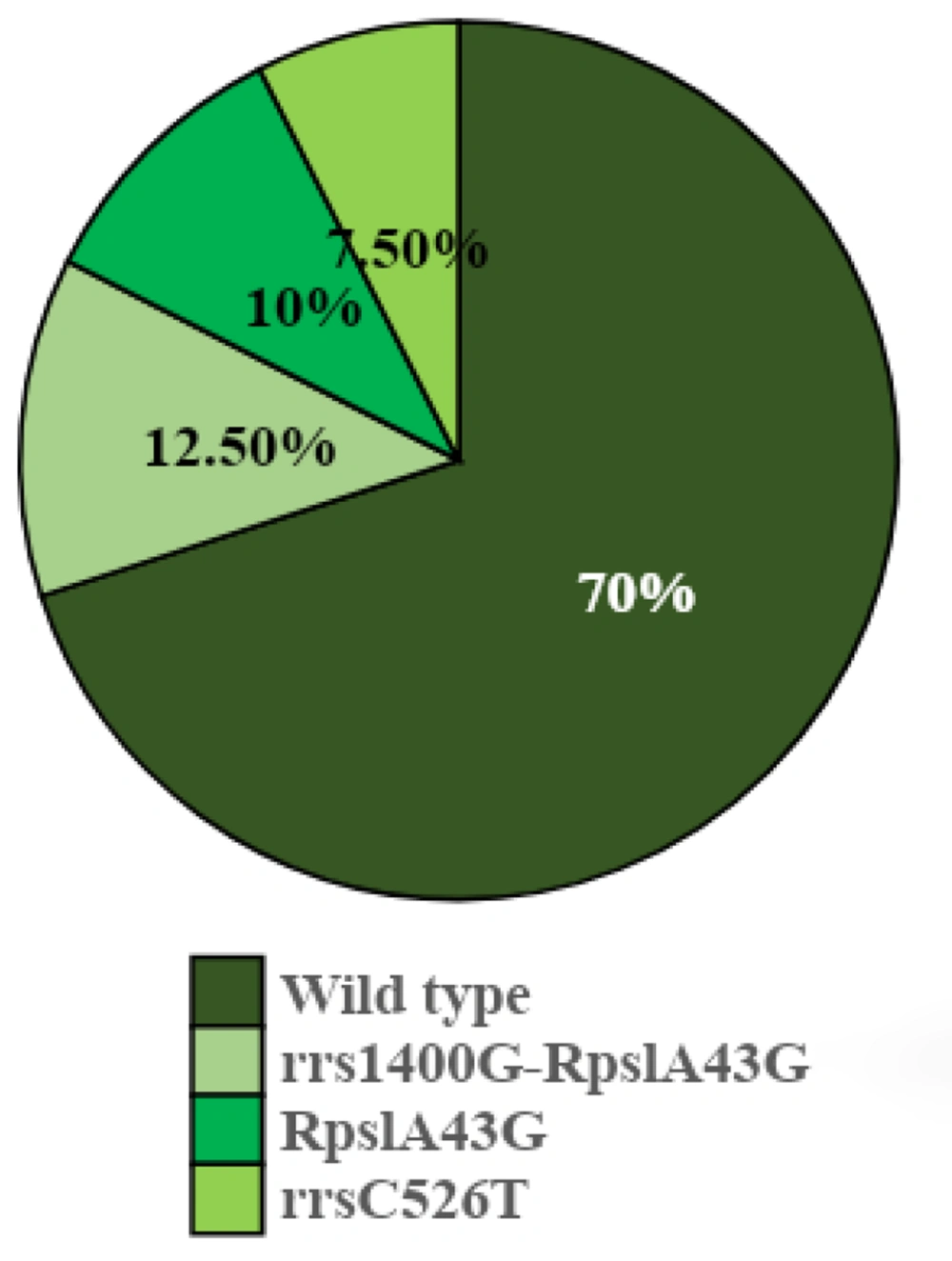
Five samples out of the 40 specimens tested have the rrs1400G-rpsLA43G mutation, and the frequency was reported to be 12.5%. Four samples showed mutations in the rpsLA43G genes, with a frequency of 10%. In 3 samples, with a frequency of 7.5% mutation, the rrsC526T gene was observed. About 28 out of the 40 specimens (frequency of 70%) didn’t show any mutations in these genes. The frequency of the analogous strains was shown in a graph of Figure 3. After hybridization, the genetic status of each bacillus was determined on the strips by staining (Figure 4).
Test results of hybridization and similar mutations. Strip No.1 bar 1, bar No.2 represents a drug-sensitive strain, strip No 3 has mutations in the streptomycin portion of the codon C526T. The number 4 strip has a mutation in the streptomycin section in codon A43G, and strip number 5 shows mutations in the kanamycin/amikacin site in codon A1400G and mutations in streptomycin in codon A43G.
4. Discussion
For regions suffering from a high tuberculosis (TB) population load such as Iran, ongoing supervision and continuous screening of drug resistance among tuberculosis based on standard drug susceptibility testing (DST) is essential for estimating the dimensions and trends of resistance to anti-tuberculosis drugs (11). In this study, substitutions that occurred in rrs and rpsL genes in drug-resistant and sensitive isolates were identified, and then comparison between strains was performed. Alterations in codons 43 (AAG → AGG) and 88 (AAA → AAG) were recorded as the main common mutations of the rpsL gene worldwide. In the current investigation, 40 specimens were used to evaluate the prevalence of point mutations related to Mycobacterium tuberculosis in Kermanshah in 2018-2019. Seventeen resistant specimens and 23 susceptible samples were studied. The frequency of mutation in strains with the lowest mutation rate was related to resistance to the rrs C526T gene mutation of Streptomycin (frequency of 7.5%), and the highest mutation rate was related to mutation in the rpsL A43G gene (frequency of 22.5%). Point mutations that occurred in other genes were also determined (frequency of 12.5%). In a study conducted by Miho Fukuda in 1999, the isolates were sequenced by researchers. About 26/121 of isolated strains were reported as resistant to streptomycin, with 78% of mutations detected in codon 43 and 22% in codon 88 of the rpsL gene (13). In 2014, a research study designed by Ryan, implemented the sequencing method and showed rpsL and rrs genes are related to TB disease and resistance of Mycobacterium to Streptomycin; after the analysis of data, it was found that in sixty percent of cases, the rpsL gene and in 25% of isolates, the rrs gene are undertaking the resistance mechanisms. According to previous studies carried out as an alternative in rpsL and rrs genes, their results revealed that rrs (8 - 15%) and rpsL (52 - 57%) had mutations, with the most frequent codon being 43 (Lys-Arg) (14).
During another investigation in 2004 conducted by WU Xue Qiong, the sequencing method was used as an evaluation method of 98 strains. The findings showed that 78/98 (80%) mutations occurred in codon 43 or 88 of the rpsL gene (Lys> Arg), 6 (6.1%) mutations occurred in the rrs gene (C-T 516) or (A-C 513), and 14 (14.3%) had no detectable alterations (15).
In 2005, Ozturk conducted a study on molecular analysis to investigate resistance against isoniazid, rifampin, and streptomycin. The sequencing method was implemented to examine 52 specimens, and the functions of five isolates were associated with streptomycin. All specimens exhibited mutations in codon 43, indicating that alterations in rpsL and rrs accounted for over 70% of resistance to streptomycin, which concurs with our findings (16).
Clinical specimens of Mycobacterium that showed mutations were found to be resistant to isoniazid, rifampin, and streptomycin according to Shenai report. Codon 43 in the rpsL gene accounted for 75% of the identified mutations in the samples. The PCR multiplex method and sequencing procedure revealed that the mutations in the rpsL gene could be identified in codons 43 and 88, and the MDR-TB isolates were found to have a significant prevalence of mutations at rpob 531, katG 315, and rpsL 43. The most considerable and common mutation detected in rpsL Lys43Arg with a 75% frequency and rrs gene 12%. About 2% of isolated specimens showed no mutations and six percent reported mutation in position 516. About 98% showed phenotype DST compliance (17).
Bifani analyzed 26 isolates by the sequencing method and 14 isolates had high resistance to streptomycin, and six samples showed low resistance. The analysis revealed that all isolated specimens possessed mutations in codon 43 of the rpsL gene and alteration consisted of lysine to threonine conversion. No changes were detected in the rrs gene (18).
In 2009, Chaoui discovered 58 Streptomycin-resistant isolates through sequencing. The study revealed that the majority of mutations were located in codon 43 of the rpsL gene, while codon 512 of the rrs gene had only a few alterations. Additionally, the research identified that codon 43 of the rpsL gene had the highest occurrence of mutations (19).
In 2012, Silke Feuerriegel investigated 97 strains using a sequencing procedure to identify mutations in the rpsL and rrs genes. The investigation discovered that the isolates which were resistant to streptomycin did not have a mutation in the rrs gene. However, 47.5% of the isolates had mutations in the rpsL gene that caused resistance to streptomycin (20).
About 90 - 95% of mutations in strains resistant to streptomycin had at least one alteration in rpsL and rrs genes and mostly occurred in bp 43 - 88 bp. The most considerable mutation in rpsL in codon 43 was accompanied by a halt in the synthesis of bacterial protein. Its subsequences are critical and have vigorous effects on DNA sequencing. Susceptible isolates didn’t show any mutation and 84.2% of isolates showed resistance to Streptomycin with mutations in rrs and rpsL genes. Consistent with other studies, rpsL has a vital role rather than rrs in resistant to Streptomycin. Findings in this study are in agreement with results obtained in our study (21, 22).
In a study by Alangaden, 1998, mutations in codon 1401 of rrs gene caused high resistance to amikacin and kanamycin in Mycobacterium tuberculosis. In his study, displacement of A to G in 13 strains was observed, that half of them were also resistant to kanamycin (23).
PCR-based methods are fast methods for detecting mutations that lead to drug resistance, but since multiple mutations in different genes may occur, all of them should be identified and require multiple PCR reactions. In our study, resistance to drugs that are common second-line drugs was observed. It is a risk factor for the treatment of patients with multiple Mycobacterium tuberculosis, and it will be difficult to treat them if the rate of multi-drug resistant bacteria is high. The resistances to these drugs lead to failure in the treatment of tuberculosis, resulting in multi-drug resistant Mycobacterium tuberculosis strains.