1. Context
Atopic dermatitis (AD) is a chronic relapsing inflammatory skin disease. It is commonly diagnosed in infancy and early childhood, with a higher prevalence than in adults, approximately 15% to 20%. In most children, it occurs within the first year of life, usually before age five. It can also be detected in children with other comorbidities of asthma and allergic rhinitis (1). Various clinical presentations are reported for AD, which might highly depend on the patient's age and disease severity. Pruritis and red, scaly, and crusted lesions on the extensor surfaces, cheeks, or scalp are typical AD presentations in infants and young children. However, it is usually characterized by more localized and lichenified plaques in older children's and adolescents' antecubital and popliteal flexures and necks (2). Functional defects in skin barriers, genetic predisposition, and immune system dysregulation mainly cause this fluctuating skin disease. Reduced levels of ceramides, filaggrin, and antimicrobial peptides (AMP), climate, air pollution, food allergies, and obesity are proposed as the main risk factors of AD (3).
There is growing evidence of microbiota's involvement in AD's pathogenesis due to its complex interaction with the local host immune system. Staphylococcus aureus is a major component of the natural human microbiota, colonizing the anterior nares of 15 – 25% of humans (4). Staphylococcus aureus colonization in the early months of age might lead to subsequent AD. Evaluating the distribution of S. aureus isolates is a matter of deep interest. Staphylococcus aureus, followed by coagulase-negative staphylococci (CoNS), is proposed as the main microbiota colonizing the skin that precedes the disease by dysregulating skin microbiota hemostasis and reducing beneficial commensal microbes.
Microbiota colonization might have pathogenetic effects through several mechanisms, including stimulation of mast-cell degranulation, the induction of keratinocyte apoptosis, stimulation of T cells, and the modulation of inflammation (5). The alternation of the nasal microorganism's composition also affects the complex pathogenesis of AD. It has been proposed that patients with AD are more frequently colonized with S. aureus than healthy controls. According to the previous studies, there are marked abnormalities between the frequency and composition of the lesional skin, non-lesional skin, and nasal microbiota in AD patients compared to healthy individuals (6). In a systematic review and meta-analysis of the prevalence of S. aureus in AD patients, almost 62% showed nasal colonization of this germ (7). They also confirmed the association of skin microbiota colonization with disease severity and age; however, the risk factors of nasal microbiota colonization are still unknown (7).
The frequency of microbiota colonization in AD patients is reported inconsistently among the studies due to the sample size, the examined patients, and different collection and detection methods (8-12). Evaluating the prevalence of nasal microbiota colonization in AD patients might reveal the important role of nasal germs colonization in disease progression and severity.
2. Objectives
In the current systematic review, we aimed to review and estimate the prevalence and odds of nasal colonization in pediatric patients with AD.
3. Data Sources
This systematic review and meta-analysis study was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement, which describes an evidence-based minimum set of items for reporting in systematic reviews and meta-analyses (www.prisma-statement.org) (13).
3.1. Eligibility Criteria
We included all types of studies (experimental and observational) on nasal colonization of children (age ≤ 19 years) with a diagnosis of AD confirmed by a physician. We imposed no language and date restrictions on the search.
3.2. Search Strategy
A comprehensive electronic literature search of PubMed and Scopus was conducted to identify relevant studies on nasal colonization in pediatric AD patients. The search was updated on 27-Jan-23. A cross-reference check was performed to expand our search and identify further relevant studies. The following search strategy was used in the databases: Atopic dermatitis AND (nasal OR nares).
4. Study Selection
Studies or subsets of studies investigating the microbiota colonization in the nasal mucosa of pediatric patients diagnosed with AD were included in the current systematic review and meta-analysis. Irrelevant studies, case reports, review articles, editorials, letters, comments, and conference proceedings were excluded. The one with a larger sample size and more complete data was selected among studies with overlapping patient data. Studies with insufficient data on nasal colonization were not included. The titles and abstracts of the studies retrieved during the initial search of databases were screened by two investigators to select relevant articles based on the inclusion and exclusion criteria mentioned. They independently screened the full texts of the remaining studies to select the eligible ones for inclusion in the systematic review and meta-analysis. Disagreements were resolved in a consensus meeting. All eligible studies with sufficient data on nasal colonization of pediatric AD patients were included in the meta-analysis.
5. Data Extraction
The following data were extracted: (1) basic study data: Author, publication year, origin country, and study design, (2) patients' characteristics: Number of patients, the detected nasal microbiota, sex, and age; the same data were extracted for the control group in case-control studies, (3) the number of patients with positive nasal germs in cohort studies and the number of healthy controls with positive nasal microbiota in case-control studies to evaluate the prevalence and odds of the nasal microbiota as the main study outcome, and (4) data regarding AD severity in patients with positive nasal microbiota to evaluate the association between nasal colonization and disease severity as the secondary outcome.
5.1. Quality Assessment
The quality assessment was conducted independently by two authors. A consensus was reached between the two authors in case of disagreements. The methodological quality of the included studies (cohort and case-control studies) was evaluated using the Newcastle–Ottawa Scale (NOS) for cross-sectional and case-control studies (14). This checklist has three major parts regarding the selection of cases and controls, comparability, and the ascertainment of the exposure.
5.2. Data Synthesis
A random-effects model was used to pool the data on the prevalence of the nasal microbiota in pediatric AD patients. In case-control studies, the prevalence of nasal colonization in patients and controls was compared and expressed as an odds ratio (OR). In cohort studies, the OR of nasal colonization was also compared with the OR of lesional and non-lesional colonization of S. aureus. Pooled data were presented with 95% confidence intervals (CIs) and obtained individually for each assay method. An I2 index was used to test for heterogeneity among studies. The I2 index is the inconsistency index and represents how much heterogeneity among the included studies is real and cannot be attributed to sampling error. Meta-regression was used when there was substantial heterogeneity between studies (I2 > 50%). Subgroup analysis was performed to compare the prevalence of nasal colonization in patients with mild, moderate, and severe AD scores (subgroup analysis and meta-regression analysis were conducted using a fixed-effects model to evaluate the predefined sources of heterogeneity).
Potential publication bias was presented using a funnel plot. The asymmetry of the funnel plot was verified using the Egger regression test (15). In the case of publication bias, Duval and Tweedle's trim and fill method was carried out to show how important publication bias can be (16). Statistical analyses were performed using Meta-MUMS DTA software (17).
6. Results
6.1. Study Selection
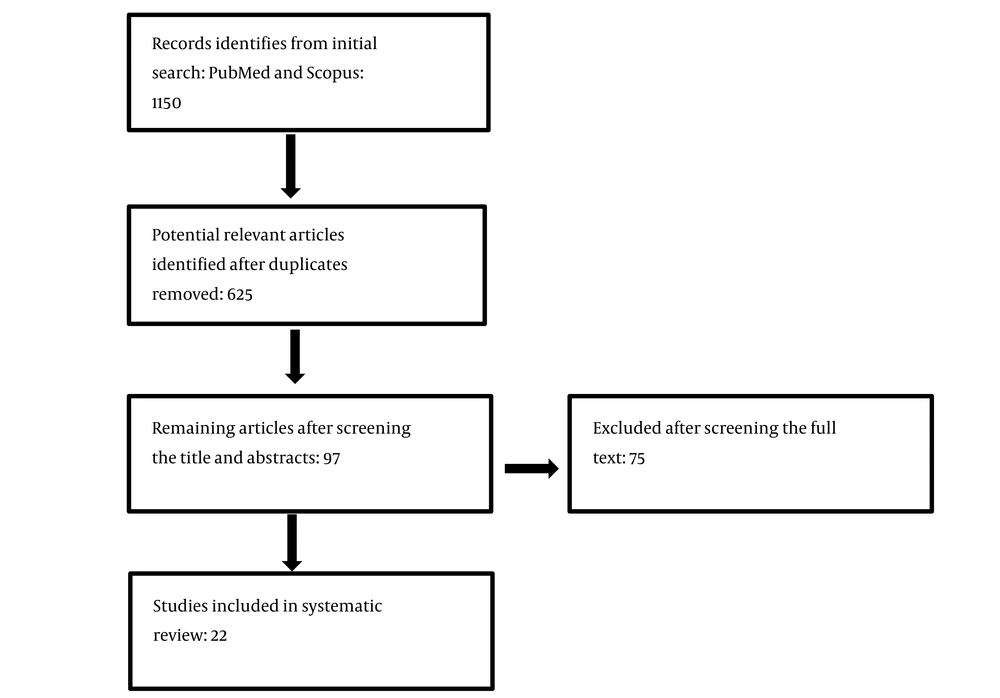
The PRISMA flow diagram of study selection is presented in Figure 1. As shown, the comprehensive electronic literature search revealed 1,150 studies. After screening the titles and abstracts and removing duplicates, 97 were selected to evaluate the full texts, and the remaining were excluded as they did not focus on the subject or were review articles, editorials, letters, comments, or conference proceedings. By reading the full texts, 22 articles met the inclusion criteria and were included in the meta-analysis. Some studies were excluded due to possible overlap in patient data (18, 19), insufficient data about the frequency of nasal colonization (20), or no access to the full text (8, 9, 21).
6.2. Study Characteristics
The main clinical characteristics of the studies are summarized in Table 1.
| Number | Study ID | Ref | Staphylococcus aureus / Total | AD Group with Nasal S. aureus | MRSA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| AD Group | Non-AD Group | Disease Severity | +/Total | |||||||
| Nasal | Lesional Skin | Non-lesional Skin | Nasal | Mild | Moderate | Severe | ||||
| 19 | Ndhlovu et al., 2022, South Africa | (22) | 29/105 | 44/104 | 33/103 | 12/87 | ||||
| 9 | Van Mierlo et al., 2022, Netherland | (23) | 8/77 | 53/68 | 24/55 | 9 | ||||
| 15 | Guimarães et al., 2022, Brazil | (24) | 29/30 | 30/30 | 24/30 | 7/12 | 6 | 20 | ||
| 1 | Abad et al., 2020, Brazil | (25) | 97/117 | 8/106 | 46 | 37 | 16 | 26/97 | ||
| 11 | Narayan et al., 2019, India | (12) | 15/55 | 16/55 | ||||||
| 8 | Wróbel et al., 2018, Poland | (11) | 23/32 | 20/32 | 9/32 | 8 | ||||
| 14 | Berbegal et al., 2018, Spain | (26) | 51/157 | 75/314 | 25 | 17 | 9 | 8/49 | ||
| 13 | Pikina et al., 2016, Russian | (10) | 30/38 | 33/38 | ||||||
| 3 | Gilaberte et al., 2015, Spain | (27) | 27/85 | 39/113 | 20 | |||||
| 7 | Jagadeesan et al., 2014, India | (28) | 92/119 | 95/119 | 18 | 57 | 5 | 24/92 | ||
| 20 | Pascolini et al., 2011, Italy | (29) | 66/119 | - | - | 18/90 | 3/9 | |||
| 17 | Kim et al., 2012, Korea | (30) | 20/44 | 18/44 | 9/44 | 7/36 | 1/20 | |||
| 2 | Balma-Mena et al., 2011, Canada | (31) | 96/200 | 87/199 | ||||||
| 4 | Chemello et al., 2011, Brazil | (32) | 28/79 | 15/79 | ||||||
| 18 | Lo et al., 2010, Taiwan | (19) | 67/133 | - | - | 170/490 | ||||
| 12 | Lebon et al., 2009, Germany | (33) | 58/209 | |||||||
| 6 | Hon et al., 2005, China | (34) | 21/55 | 16/20 | 1/32 | |||||
| 21 | William et al., 1999, USA | (35) | 54/100 | 78/100 | - | 43/100 | ||||
| 22 | Dhar et al., 1992, India | (36) | 17/50 | 25/50 | 13/50 | 7/50 | ||||
| 16 | Hoeger et al., 1992, Germany | (37) | 35/41 | 37/41 | 30/41 | 10/41 | ||||
| 5 | Falanga et al., 1985, USA | (38) | 11/12 | 11/12 | ||||||
| 10 | Smith et al., 1975, England | (39) | 11/20 | 18/20 | ||||||
All the selected studies were observational, with sample sizes ranging from 12 to 209. In nine of the studies, the odds of S. aureus nasal colonization were compared between AD pediatrics and non-AD healthy controls (19, 22, 24, 26, 29, 30, 35-37), and the 13 remaining studies included only AD cases with no control group (10-12, 23, 25, 27, 28, 31-34, 38, 39). There was no considerable difference in the number of AD males and females (a slight male preponderance was noted), and cases were under 19 years old. The swabbing procedure was used for anterior nares and skin colonization evaluation.
The Scoring Atopic Dermatitis (SCORAD) index and EASI scoring system determined the disease severity.
6.3. Prevalence of Staphylococcus aureus Nasal Colonization
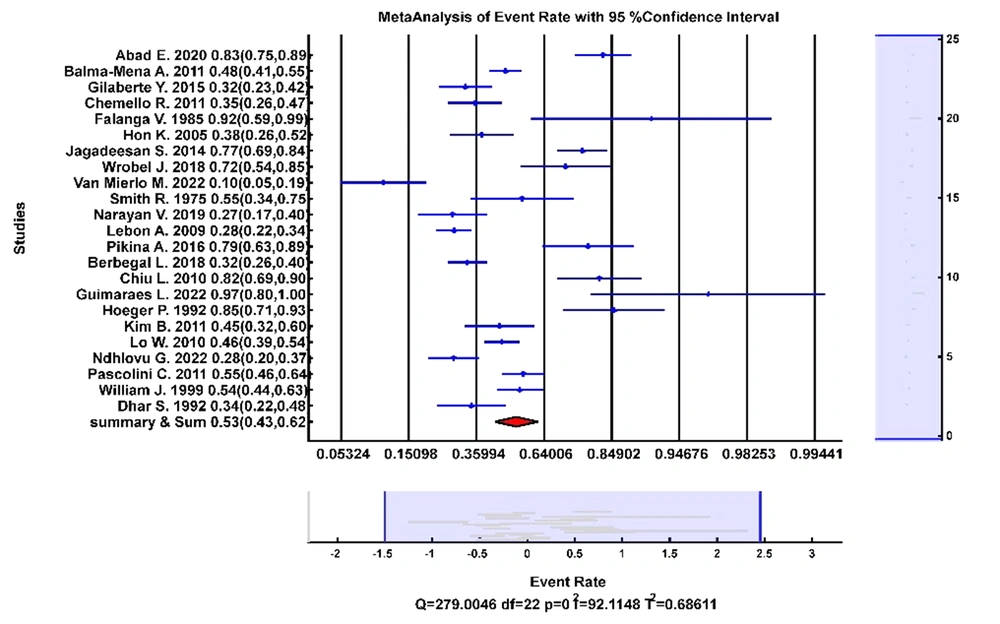
Thirteen studies had enough information to synthesize the prevalence of S. aureus nasal colonization, which included 1,960 AD pediatrics. Figure 2 shows the forest plot of the prevalence rates. Pooled analysis showed that the prevalence of S. aureus was 53% (95% CI: 0.43 - 0.62) in AD pediatric noses (P < 0.001, Q = 307, I2 = 92.84%). The studies had considerable heterogeneity, possibly due to patient characteristics and disease severity variations.
Subgroup analysis was performed on eight studies with data regarding the AD severity in pediatrics with positive nasal colonization of S. aureus. Pooled analysis showed that S. aureus was colonized in 38% of the pediatrics with mild AD, 50% with moderate AD, and 22% with severe AD.
6.4. Odds of Nasal Colonization with Staphylococcus aureus
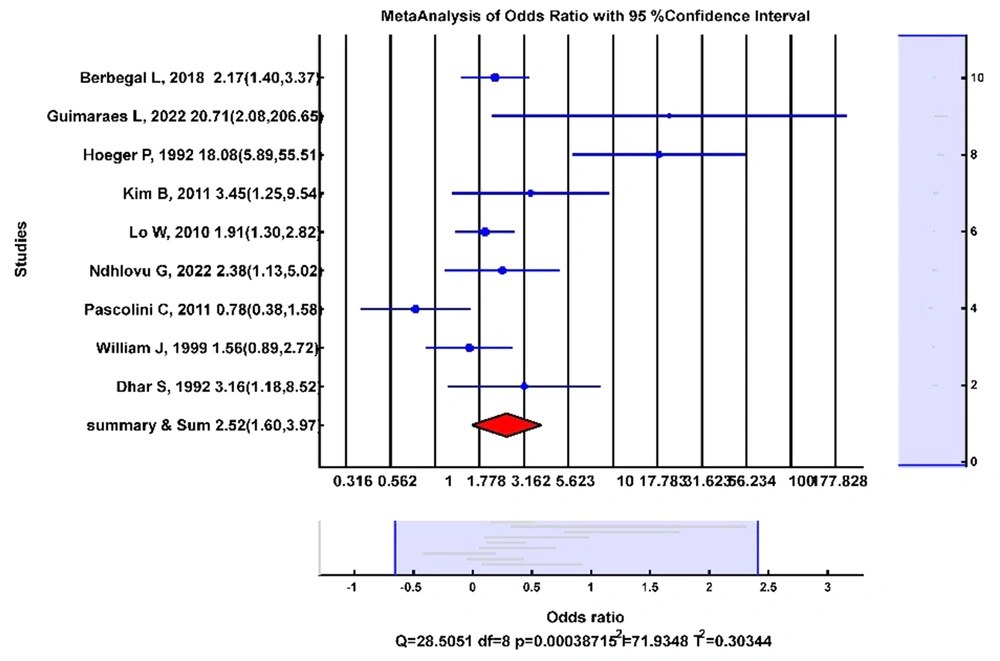
Nine studies had data on the odds of S. aureus in AD pediatric noses compared with non-AD healthy controls. Figure 3 shows the forest plot of the OR analysis. The random-effects model showed that the odds of nasal colonization of S. aureus were significantly higher in AD pediatrics than in non-AD healthy controls (OR: 2.52; 95% CI (1.60, 3.97); I2 = 72%).
According to 17 studies, the odds of S. aureus were lower in the noses of AD pediatrics than in their lesional skin (OR: 0.68; 95%CI (0.35 - 1.33); I2= 90%). Five studies showed that the odds of S. aureus were higher in the noses of AD pediatrics than in their non-lesional skin; however, it was not statistically significant (OR: 2.02; 95% CI (0.89, 4.51); I2 = 83.3%).
Meta-regression was performed on six studies regarding the mean age of patients with positive S. aureus colonization to explain statistical heterogeneity in the associations between age and nasal colonization. Based on the results, meta-regression showed that age was not a source of heterogeneity.
The prevalence of MRSA among AD children with S. aureus nasal colonization was 24%. The odds of MRSA in AD children with S. aureus nasal colonization compared with non-AD healthy controls showed a pooled OR of 1.41 (95%CI (0.57, 3.5)).
6.5. Other Nose Microbiota Colonization
Two studies reported data on other staphylococcal species besides S. aureus in the nose of AD pediatrics (22, 24). The pooled data revealed a prevalence of 5% for S. capitis, 32% for S. epidermis, 4% for S. haemolyticus, 4% for S. hominis, 8% for S. saprophyticus, and 2% for coagulase-negative staphylococcal spp in the anterior nares of AD children (22, 24).
6.6. Publication Bias
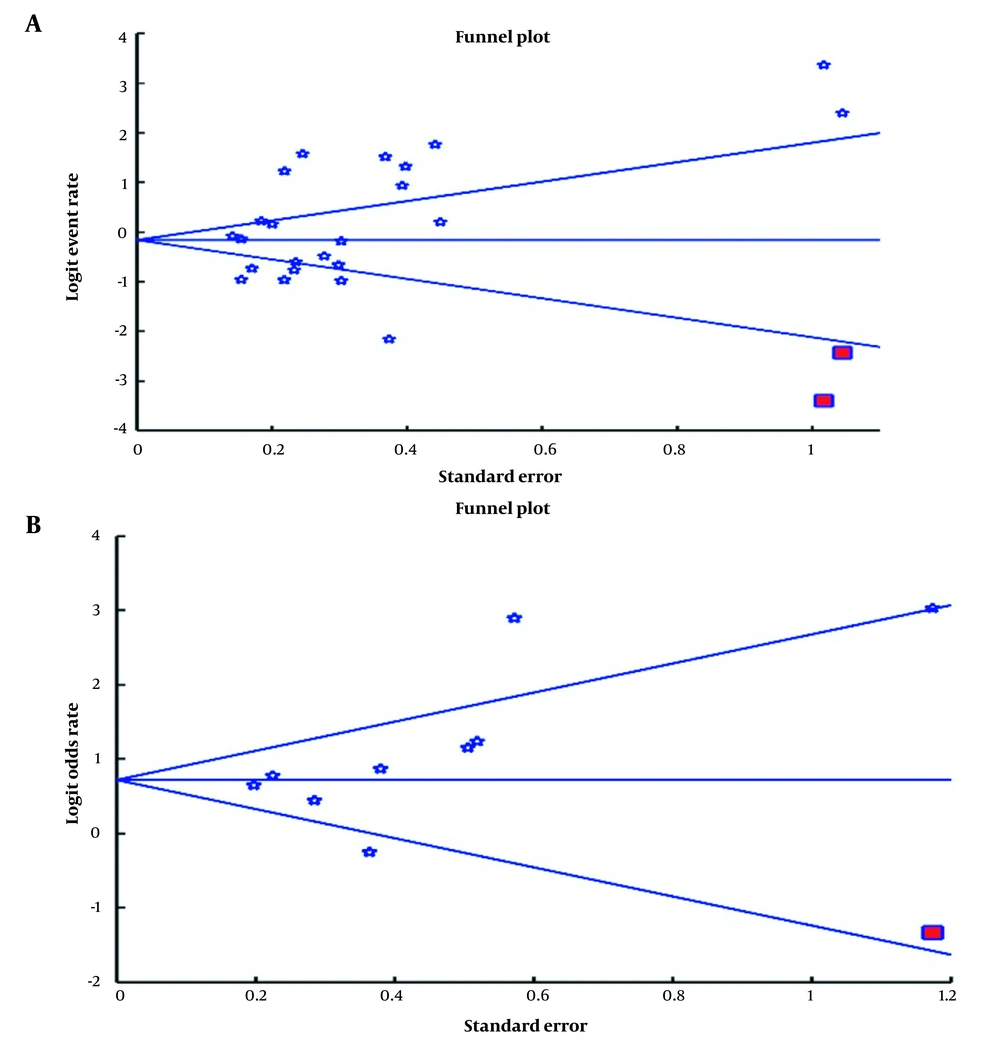
Publication bias was used to examine bias in the reported results. The funnel plot of the studies on S. aureus prevalence rate (A) and odds ratio (B) seemed asymmetric (Figure 4A and B). The asymmetry of the funnel plots was checked using Egger's regression intercepts and revealed no significant publication bias regarding the prevalence of S. aureus and the odds of S. aureus in the nose of AD pediatrics compared with non-AD healthy controls. Furthermore, we applied the trim-and-fill method to this meta‐analysis, which did not change the results in the primary analysis after trimming two studies. The publication bias did not influence the significance of our results.
6.7. Quality Assessment of the Studies
The quality of controlled and uncontrolled studies was evaluated with the Newcastle-Ottawa Scale. Detailed data regarding the selection of cases and controls, comparability, and ascertainment of the exposure were evaluated and presented in Table 2. According to Table 2, there were some studies with low quality and insufficient data regarding the case selection and severity of AD in cases with nasal S. aureus colonization.
| Studies with the Case Group | ||||||||
|---|---|---|---|---|---|---|---|---|
| Study ID | Ref | Representativeness of the Exposure Group | Diagnosis of AD | Disease Severity | Selection of Control | Comparability | Exposure Ascertainment | Assessment of the Outcome |
| Van Mierlo et al., Netherland, 2022 | (23) | Difficult-to-treat AD | Not reported | SA-EASI | - | - | Swabbing method | DNA isolation, qPCR, and sequencing |
| Abad et al., Brazil, 2020 | (25) | Hospital-based | Not reported | SCORAD | - | - | Swabbing | Culturing and PCR |
| Narayan et al., India, 2019 | (12) | - | Hanifin and Rajka | SCORAD | - | - | Swabbing method | Culturing methods |
| Wróbel et al., Poland, 2018 | (11) | Consecutive | Hanifin and Rajka | - | - | Swabbing method | Culture on solid and broth media | |
| Pikina et al., Russian, 2016 | (10) | - | Not reported | - | - | Swabbing method | Culturing method | |
| Gilaberte et al., Spain, 2015 | (27) | Consecutively | Hanifin and Rajka | - | - | Swabbing | Culture | |
| Jagadeesan et al., India, 2014 | (28) | Consecutive/outpatient clinics | Hanifin and Rajka | EASI | - | - | Swabbing method | Qualitative bacterial culture |
| Balma-Mena et al., Canada, 2011 | (31) | Dermatology Clinic | By pediatric dermatologists | AD severity score (0–25) was designed | - | - | Swabbing | PCR |
| Chemello et al., Brazil, 2011 | (32) | Sanitary Dermatology Outpatient Clinic | By the main researcher (dermatologist) | Not reported | - | - | Sterile swabs | Culturing methods |
| Lebon et al., Germany, 2009 | (33) | The Generation R Study Center | Not reported | Questions that pertain to the level of suffering | - | - | Swabbing method | Culturing methods |
| Hon et al., China, 2005 | (34) | - | Managed at a pediatric dermatology service | - | - | Swabbing method | Bacterial culture of swabs | |
| Falanga et al., USA, 1985 | (38) | Consecutive AD patients | Not reported | - | - | Swabbing method | Quantitative cultures for Staphylococcus aureus | |
| Smith et al., England, 1975 | (39) | - | Not reported | - | - | Swabbing method | Culturing methods | |
| Case-control Studies | ||||||||
| Case-control studies | Ref | Representativeness of the Exposure Group | Diagnosis of AD | Disease Severity | Selection of Controls | Comparability | Exposure Ascertainment | Assessment of the Outcome |
| Guimarães et al., Brazil, 2022 | (24) | - | Hanifin and Rajka's criteria | Non-AD sibling of the children | Severity | Sterile swab | MALDI-TOF-MS | |
| Ndhlovu et al., South Africa, 2022 | (22) | Hospital-based | Based on the United Kingdom Working Party diagnosis of atopic eczema | SCORAD | Non-AD from community early development centers | Age | Swabbing | Culturing |
| Berbegal et al., Spain, 2018 | (26) | Consecutive/Pediatric Dermatology Clinic | Diagnostic criteria of the consensus conference on pediatric atopic dermatitis | SCORAD | Clinic children with no AD or dermatological condition | Age/severity | Sterile swab | Culturing methods |
| Kim et al., Korea, 2012 | (30) | Four kindergartens | Hanifin and Rajka | EASI | Healthy volunteers | Not reported | Sterile swab | Culturing and PCR |
| Lo et al., Taiwan, 2010 | (19) | Pediatric Department | Hanifin and Rajka | Not reported | Healthy children in the community | Age and sex-matched | Swabbing | Culturing and PCR |
| Pascolini et al., Italy, 2011 | (29) | Clinic-based | Hanifin and Rajka | SCORAD | Healthy children | Age/sex | Swabbing | Culturing and PCR |
| William et al., USA, 1999 | (35) | Hospital AD children | Not reported | Rajika and Langeland | Children with non-AD dermatologic conditions | Age/sex | Swab | Culturing |
| Dhar et al., India, 1992 | (36) | AD patients | Not available | Not available | Healthy children | Age/sex | Swab | Culturing |
| Hoeger et al., Germany, 1992 | (37) | - | Standard criteria | Children at an outpatient clinic for assessment of cardiac function | - | Sterile swab | Culturing method | |
7. Discussion
The current systematic review and meta-analysis pooled the data from all the available published literature on nasal colonization of pediatric patients with AD. Pooled estimate revealed a high prevalence of S. aureus in AD pediatric noses (51%), consistent with our expectations. Although S. aureus colonizes the anterior nares of some healthy individuals asymptomatically and permanently, we indicated that it would be considerably colonized higher in the nasal of AD pediatrics than in healthy children's noses, which is particularly important. The significantly higher incidence rate of nasal colonization indicates the involvement of S. aureus nasal colonization in the pathogenesis of AD in pediatrics.
According to our results, the possibility of nasal colonization of S. aureus was 2.5 higher in AD pediatrics than in healthy children. We evaluated only studies on pediatric patients with AD in the current meta-analysis, and the obtained results confirmed a previous meta-analysis performed in 2016 on the prevalence of S. aureus in the skin and nose of AD patients of different ages (7). They proposed a 57% prevalence of S. aureus colonization in AD patients. According to our results, although the odds of S. aureus colonization were significantly higher in the anterior nares of pediatric AD patients than in healthy controls, the noses of AD children had a lower S. aureus colonization rate than their lesional skin. However, it was more prevalent in the anterior nares of AD pediatrics than in their non-lesional skin. Similarly, higher S. aureus colonization was detected in the patients' skin than in their noses by Totte et al. (7). It could be concluded that concomitant colonization of toxigenic S. aureus is considerable in the nose and skin of AD pediatrics. Several studies also reported indistinguishable isolates and a genetic relationship between S. aureus isolates colonizing noses and the skin (29, 37).
There is a lack of knowledge on the immune response that controls nasal colonization; however, bacterial adhesion, defective bacterial clearance, and decreased innate immune response might be involved in the increased colonization of S. aureus in AD children compared with healthy cases (40). Pooled data from five studies showed that the prevalence of MRSA was 24% in pediatrics with nasal colonization of S. aureus. According to Jagadeesan et al. study on Indian pediatrics with AD, 25 out of 92 had MRSA nasal colonization, being the highest reported prevalence of MRSA in AD pediatrics among other included studies. They suggested an association between the proportion of nasal colonization with MRSA and the geographical area (28). The other reason for the higher prevalence of MRSA in some studies might be related to outpatient settings compared with hospital-based settings. Community-acquired MRSA (CA-MRSA) strains could be differentiated from hospital-associated MRSA (HA-MRSA) regarding genetic, epidemiologic, or microbiological profiles. Patients with AD are not commonly admitted to the hospital, making them more likely to become colonized or infected with CA-MRSA. In the study of Jagadeesan et al., the higher prevalence of MRSA in outpatient settings was related to the presence of the MRSA gene alone in CA-MRSA strains compared with multiple antibiotic resistance in healthcare-associated MRSA (28).
Based on our findings, there was no significant association between S. aureus nasal colonization and the mean age of the pediatrics with AD, which might be due to the low number of studies with sufficient data regarding the mean age of pediatrics with S. aureus nasal colonization. Our finding was consistent with other studies that could not confirm any association between the patient's age and the prevalence of S. aureus in AD patients' noses and non-lesional skin (7). The main results might be the low sample size of the studies, considerable heterogeneity among the included studies, and low quality of the studies on nasal colonization of S. aureus in children with AD. Pediatric patients with AD were the target population of the included studies in the current review, while children in early life are not considerably exposed to risk factors of bacterial colonization, such as colonized people and environment (41), or might have immature microbiota. So, there are still uncertain data regarding the age distribution of S. aureus nasal colonization in children. Berbggal et al. showed a higher prevalence of nasal colonization in children with AD at age six or older than in younger children (26).
There is also no consensus on the disease severity among AD patients with S. aureus colonization and those without. In some studies, nasal colonization of S. aureus has been associated with more severe symptoms of the disease and skin infection; however, no relationship has been detected by others (4, 34, 42). By pooling the data of the studies on AD pediatrics, we obtained a significant relationship between increased severity and prevalence of S. aureus. According to the current study, S. aureus nasal colonization was high in children with moderate AD, and S. aureus colonization was less prevalent in the noses of pediatrics with mild AD. However, children with severe AD showed lower nasal colonization of S. aureus than those with moderate AD. This might be related to methodological variation between studies and the low number of children with severe AD in studies to get statistically significant differences. Some previous studies support our findings by comparing the relationship between AD severity and S. aureus prevalence between noses and skin. They showed a higher prevalence of S. aureus colonization in the skin of patients with severe AD; however, they did not find any association between nasal colonization of S. aureus and the severity of AD (27, 42, 43). It has been suggested that the skin microbiome correlates more with disease severity than the nasal microbiome (20). Pediatric AD patients with mild or moderate AD and S. aureus nasal colonization might reveal more severe symptoms throughout childhood. On the other hand, the absence of severe or systemic infections in most AD patients could be related to the enhanced oxidative metabolism of polymorphonuclear leukocytes or increased cell-mediated immune responses to staphylococcal antigens in AD (31).
The role of nasal S. aureus in AD is controversial and a crucial research area. Anterior nares (vestibulum nasi) is one of the most frequent sites and main reservoirs of gram-positive S. aureus in children with AD, which is involved in self-contamination, the spread of the pathogen, and the transmission of S. aureus from the nose to the skin at early childhood (44). Also, S. aureus colonization in AD might be a potential risk factor for different invasive conditions such as bacteremia, septic shock, osteomyelitis, necrotizing pneumonia, or septic arthritis, so S. aureus colonization in AD children should be detected and eradicated as early as possible (45).
Despite studies on S. aureus, its pathologic role in AD is still under investigation, and the involvement of other staphylococcal species is poorly understood. Only two studies with sufficient data on other staphylococcal species were detected in AD children's noses. Pooled data showed that the most isolated species was S. epidermidis, as previously reported by other studies on skin colonization of adult AD patients (46, 47). The bacterium colonization rate might be related to its protective or pathogenic role in disease severity. Similar to our results, S. hominis had a low prevalence in several studies, possibly related to its protective role in AD (24).
As a chronic, relapsing-remitting inflammatory disease, AD can be challenging to treat, so higher nasal colonization in AD children compared with non-AD cases shown in the current systematic review and meta-analysis could be promising for promoting the quality of life and intensity of the disease during the treatment period in AD pediatrics through eradicating nasal cavity S. aureus.
7.1. Limitations
There was some limitation in our systematic review. First, the prevalence of S. aureus was a secondary objective in some of the included studies, so data regarding the characteristics (age, sex, and disease severity) of children with nasal colonization were not presented in detail by these studies. Second, due to the low sample size of children with severe AD in the included studies compared with cases with mild and moderate AD, we could not prove the exact role of S. aureus colonization in the severity of AD. Accurate evaluation of the association between S. aureus colonization and AD severity needs more studies on children with severe AD. Studies with large sample sizes and high quality on the prevalence of S. aureus in the nose of AD children versus healthy controls might increase the validity of the pooled estimate.
7.2. Conclusions
Pooled data on nasal microbiota colonization in pediatrics with AD showed the higher prevalence of S. aureus colonization in AD children compared with non-AD healthy controls, confirming the pathophysiologic role of S. aureus colonization in AD. Although children with moderate AD showed higher nasal colonization of S. aureus than cases with mild AD, this study could not prove the role of Staphylococcus colonization in the severity of atopic dermatitis. More studies on children with severe AD are needed.