1. Background
Group B streptococcus (GBS; Streptococcus agalactiae) is a facultative aerobic Gram-positive organism with a structure similar to other Gram-positive cocci. This bacterium is a part of many females' vagina and gastrointestinal tract normal flora (commensal organism); but, it can cause life threatening infection in susceptible hosts, including newborns, pregnant/postpartum women, and also adults with underlying diseases such as diabetes and malignancy. In fact, GBS has emerged as a notable cause of human disease during several decades and nowadays it is the most common cause of life threatening invasive bacterial infections (septicemia, pneumonia and meningitis) during the neonatal period (1-4).
About 10-30% of pregnant women have GBS colonization in vagina or rectum and thereby 50-70% of their neonates acquire GBS through the birth canal during delivery. Therefore, the GBS colonization occurring at the end of pregnancy phase is a risk factor for neonatal infection and identifying/screening of pregnant women with GBS colonization is significant in preventing neonatal GBS infection (5). The Centers for Disease Control and Prevention (CDC) has estimated the fatality rates among infants to be 4-6% in the United States and about 10% in the United Kingdom and Ireland. The neonatal GBS disease is more common than other known diseases such as rubella and spina bifida which affect newborns (5, 6).
Pregnant women may also manifest disease association with GBS including urethral tract infection (ureteritis) and/or serious diseases such as chorioamnionitis, endometritis and bacteremia (postpartum fever) during pregnancy, delivery or postpartum period (4). Furthermore, maternal infection may cause preterm delivery, low birth-weight infants or stillbirths (7). GBS colonization is asymptomatic and occurs in a transient, chronic or intermittent manner and the colonization period is variable; therefore, detecting GBS in vaginal tract at certain times of pregnancy could indicate the existence of this bacterium at delivery (8). Performing culture or tests in fewer than five weeks before delivery are relatively accurate for predicting the carrier's situation at the time of delivery (9).
Currently, the recommended gold standard method by CDC for identifying GBS rectovaginal colonization at 35-37 weeks of gestation (perinatal screening method) is based on bacteriological criterion, i.e. culturing rectovaginal swab specimens in selective broth and subsequent subculture on medium. However, it requires 24-72 hours for getting the results which is not rapid enough for GBS detection at or close to the time of delivery. Moreover, according to reported studies, bacteriological methods can predict only 87% of GBS colonization possibility in women’s rectovaginal at delivery. Several studies have shown a low sensitivity of prenatal GBS culture for identifying colonization during pregnancy or at delivery and low sensitivity and specificity of other tests (rapid antigen-based tests and hybridization-based methods) for direct detection of GBS from clinical specimens of colonized pregnant women (5, 10). Rapid screening tests for detecting GBS that can be helpful to identify carrier mothers a few days before delivery or at delivery will improve the need for screening pregnant women at 35-37 weeks of gestation, thereby avoiding from prescribing nonessential prophylactic antibiotics during delivery for women with no GBS colonization (5).
Applying rapid, accurate and sensitive method for detecting GBS and receiving intrapartum antibiotic prophylaxis (IAP) at delivery have been demonstrated to increase the treatment possibility of carrier pregnant women and decrease the rates of GBS vertical transmission to infants; in other words, rapid and reliable detection of GBS colonization will be effective for pregnant women particularly the ones who have received poor prenatal treatment during pregnancy, thereby allowing for effective prevention of neonatal and maternal GBS infection (11).
2. Objectives
The aim of this study was to use culture method as the gold standard criterion recommended by CDC in 2010 as well as polymerase chain reaction (PCR) targeting 16S rRNA primers for detection of GBS in vaginal specimens of pregnant women at 35-37 weeks of gestation in Hamedan, Iran.
3. Patients and Methods
3.1. Study Design
The study was performed from June 2013 to February 2014. The study was approved by the ethical committee of Hamadan university medical sciences. The included individuals were women at 35-37 weeks of pregnancy with no clinical problem, referred to prenatal care of Fatemieh Hospital and/or private clinic centers in Hamadan. The criterion for exclusion was being a pregnant woman at < 35 weeks of gestation. The information about age and history of antibiotic usage until two weeks prior to the study for each pregnant woman was questioned and recorded. A total of 203 double-vaginal specimens were collected using sterile cotton tip swabs from lower vagina for each pregnant woman. One of the swabs was placed into an examination tube containing Stuart transport medium (for microbiological culture) and the other one was placed into an examination tube containing 1 mL phosphate buffered saline (1x PBS) with pH = 7.2 (for direct DNA extraction and performing PCR). The specimens were transferred to the bacteriology laboratory to be processed within 24 hours.
3.2. Isolation and Detection by Culture Method
Upon the collected samples were transferred to the bacteriology laboratory, the swab specimens in Stuart transport medium were removed and inoculated into Todd-Hewitt broth (Pronadisa Co., Spain) supplemented with 1% yeast extract, 10 µg/mL of colistin and 15 µg/mL of nalidixic acid (Lim broth). This inoculated selective broth was incubated at 35-37°C in 5% CO2 (in candle jar) for 18-24 hours. The incubated broth was subcultured to a plate containing tryptic soy agar with 5% defibrinated sheep blood and was incubated at 35-37°C in 5% CO2 for 18-24 hours. Afterwards, the agar plates were inspected and identified for organisms suggestive of GBS by the following criteria: colonies with narrow zone of beta hemolysis, Gram-positive cocci, catalase negative, resistance to bacitracin and trimethoprim/sulfamethoxazole (SXT) diagnostic antibiotic discs, sodium hippurate hydrolysis-positive, and CAMP [Christie, Atkinson, Munch, Peterson] positive. Based on reported studies, approximately 5% of GBS strains have no hemolysis on blood agar; so, suspected typical GBS colonies without hemolysis were also examined. After no GBS was identified on agar plates, the suspected GBS colonies were reexamined after an extra 18-24 hours of incubation. The time required for performing the culture method was estimated about 54-72 hours for one sample.
3.3. DNA Extraction and Polymerase Chain Reaction Assay
DNA was extracted by thermal lysis (boiling method) (12) with some modifications as follows: To concentrate the nucleic acid obtained from the boiling method, 400 μL of ice-cold ethanol was added into the micro tube containing nucleic acid, it was mixed gently and left in a deep freezer (-18 to -20°C) for 10-30 minutes; then, it was centrifuged for 10 minutes at the maximum speed rpm. The supernatant was decanted and the pellet was dried out thoroughly using heater block instrument (Nedaye Fan Co., Iran) at 55°C. Finally, 50 μL of sterile 1x TE [Tris-EDTA buffer] buffer was added to the micro tube and the pellet was gently dissolved. The concentrated nucleic acid was used for PCR or was frozen at -20°C until further use.
The 405-bp 16S rRNA gene was chosen as the GBS primer (GenBank accession number: 2353759) for PCR. The forward and reverse sequences of the primers were CGCTGAGGTTTGGTGTTTACA (40-61) and CACTCCTACCAACGTTCTTC (445-465), respectively (13). Homology checking was performed using the BLAST database. The action was repeated again for the positively resulted specimens. Like wise, the three other atr (GI No. AF15135), cfb (GI No. X72754) and scpB (GI No. AF189002) genes, as previously described (10, 12, 14), were used for testing all the yielded positive results of 16S rRNA gene to verify the true positive results. These primers were synthesized by Bioneer Oligo Nucleotide Synthesis Co. (Korea).
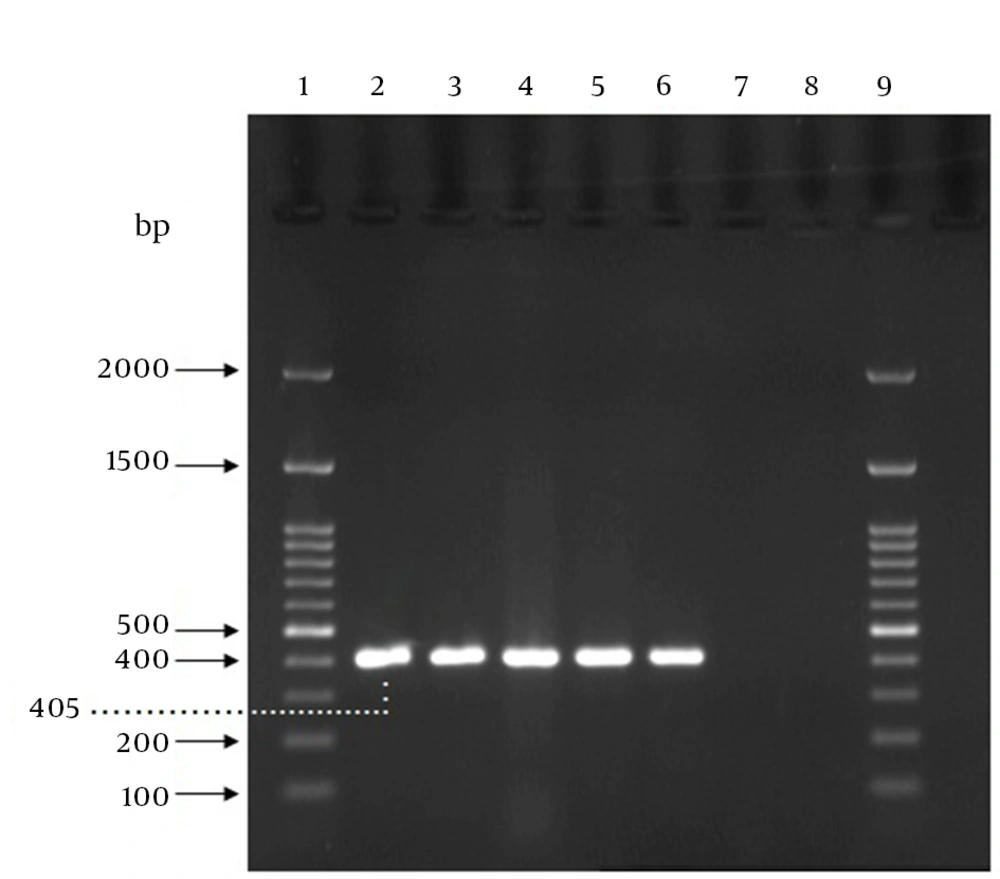
The PCR reaction was 20 μL which was prepared as follows: 10 μL of 2x premix master mix (Parstous Biotech Co., Iran), 5 μL sterile double-distilled water, 1 μL of the forward primer, 1 μL of the reverse primer, and 3 μL of the DNA sample. The DNA samples as well as a positive control (a sample including DNA from S. agalactiae ATCC 12386) and a negative control (a sample without template) were amplified by an initial denaturation step for five minutes at 94°C, followed by 35 cycles of 94°C for 45 seconds, 60°C for 60 seconds, and 72°C for one minute, and a final cycle of 72°C for seven minutes in a Bio-Rad thermal cycler. After amplification, 5 µL of each amplification product was combined with 1 µL of 6x DNA loading dye buffer (Parstous Biotech Co., Iran) and analyzed by electrophoresis on a 1.5% (w/v) agarose gel, stained with DNA safe stain (Cinna Clone Co., Iran). The 100-bp molecular size marker (Cinna Clone Co., Iran) was run concurrently. The electrophoresis was carried out in 1x Tris, boric acid, EDTA (TBE) at 80 V for one hour. The band pattern of the amplified products on the agarose gels were visualized and photographed using UV Tran illuminator (Vilbert Lourmat Co., Japan). The samples presenting a 405-bp amplicon were considered positive for GBS, as shown in Figure 1. The time required for performing direct DNA extraction and PCR assay was estimated about four hours for one sample.
Lanes 1 and 9,100 bp DNA ladder; lane 2, GBS ATCC 12386 (positive control); lanes 3, 4 and 5, positive specimens in both PCR and culture method; Lane 6, the specimen that was negative in culture method but positive by PCR; lane 7, the specimen that was positive using culture method but negative by PCR; lane 8, negative control (sample without template).
3.4. Determining Specificity and Limit of Detection (Sensitivity) in Polymerase Chain Reaction (16S rRNA Primers)
Proving no sequence homology with any genes in other species is necessary for precise detection of a certain microorganism. Thereby, to assess the specificity of the PCR assay, a battery of 10 non-GBS microorganisms (Table 1) was tested by the gene. The lower limit of detection (sensitivity) of the PCR was determined by performing a 10-fold serial dilution of 108 CFU/mL (standard 0.5 McFarland) GBS from 108 to 100 CFU/mL in nine sterile examination tubes containing 1 mL sterile physiological serum and vaginal discharge from healthy females (not infected with GBS) concomitantly. Moreover, aliquots of the nine dilution serials were added to the nine examination tubes containing 1 mLof 1x PBS using specimen swabs verified free of GBS. Genomic DNA was extracted using the method discussed above. GBS was detected from each concentration in a duplicate, using PCR.
3.5. Statistical Analysis
When a specimen was positive by culture or PCR, it was considered positive for GBS. The PCR results were compared with the culture results to determine sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of PCR for direct detection of GBS in vaginal tracts of pregnant women. Considering P value of ≤ 0.05 as statistically significant and 95% confidence interval, the correlation of age groups (≤ 30 and > 30) and antibiotic taking history with rates of positive or negative results for GBS vaginal colonization was determined by Pearson correlation analysis and chi square test (χ2). All the statistical analyses were performed by SPSS software (version 20).
4. Results
To evaluate the specificity of the primers targeting the GBS-16S rRNA gene, a panel of 10 non-GBS bacterial species was tested by PCR using the 16S rRNA gene. No product (amplification band) was detected for the seven non-GBS species; however, a 405-bp faint PCR product was visualized in the cases of S. aureus ATCC 25423, E. faecalis ATCC 29212 and E. faecium (clinical isolate), indicating that there was some homology in the 16S rRNA gene regions among GBS strain and the mentioned bacteria (Table 1).
Serial dilution studies were performed to establish the limit of detection of PCR targeting 16S rRNA. This method was reproducible with serial dilution and PCR was performed in duplicate. DNA extraction followed by PCR indicated that as few as 100 copies of GBS cells per milliliter can be detected by PCR. In fact, the limit of detection of this PCR for serially diluted GBS in the sterile physiological serum and in PBS with specimen swab verified free of GBS was 102 CFU/mL.
The prevalence of GBS vaginal colonization in the 203 obtained specimens was 7.39% (15 samples) by culture method and 19.7% (40 samples) by PCR (Table 2); 27 specimens showed positive result by PCR and negative by culture; two specimens were positive by culture and negative by PCR. All the positive results were repeated for assurance of true results. Furthermore, as shown above, to confirm the specificity of PCR for detecting GBS, all the PCR positive specimens targeting the 16S rRNA gene (more than 40 cases) were tested using three other genes including atr, cfb and scpB to verify the true positive results. Although each of these genes separately could not detect all the positive results yielded from using 16S rRNA, the three genes verified 40 positive specimens resulted from 16S rRNA in sum (data not shown). Consequently, 42 specimens were considered as true positive (27 samples only using PCR, two samples only using culture and 13 samples using both methods). Therefore, overall GBS vaginal prevalence in this study was determined 20.69%.
The statistical analysis for PCR results as a direct diagnostic test were compared with that of culture method as the gold standard criterion, which revealed a sensitivity of 88.23%, specificity of 87.44%, PPV of 35.71%, NPV of 98.94%, and accuracy of 87.50%. The mean age of all the attending individuals was 27.10 years (ranging 16-42 years). There was no significant correlation related to antibiotic taking history and age groups with the presence of GBS bacterial colonization.
| Tests | Culture (Gold Standard) | Total | |
|---|---|---|---|
| Presence of GBS (Positive) | Absence of GBS (Negative) | ||
| PCR (16S rRNA gene) | |||
| Positive | 13 | 27 | 40/203 = 19.70% |
| Negative | 2 | 161 | 163 |
| Total | 15/203 = 7.39% | 188 | 203 |
Detection Results of Group BS treptococcusin Vaginal Tracts of Pregnant Women Using Culture and PCR a
5. Discussion
Except for a future promising vaccine, one of the strategies to prevent GBS disease is screening and intrapartum antibiotic prophylaxis. Prescribing prophylactic antibiotics for GBS carrier pregnant women during delivery can decrease vertical transmission of GBS to neonates; thereby decrease early onset disease (EOD) in neonates (5). Culture method, antigen-based tests and hybridization-based methods lack enough sensitivity and these methods can only identify women with heavy GBS colonization. In addition, they are time-consuming (5, 10). Using improved diagnostic instruments such as PCR for detecting GBS may result in reasonable utilization of antibiotics. In fact, combining rapid sampling with rapid proliferation and identifying technologies can aid to improve the control and prevention of infectious diseases (11).
Performing Pearson correlation analysis, no variables related to ages were significantly associated with the presence of GBS colonization in vaginal tracts of pregnant women; (P = 0.622). Of the 203 pregnant women, 41 stated a history of antibiotic taking until two weeks prior to the study and five of them were positive (three samples using only PCR method plus two samples using both PCR and culture methods). In comparison, 162 individuals had not taken antibiotic that 37 of them were positive (24 samples using only PCR method, two samples using only culture method and 11 samples using both). However, performing Pearson correlation analysis and chi square test (χ2), no significant correlation was found between antibiotic taking history and the presence of GBS colonization (P = 0.178). Rates of colonization in 20.69% were similar to the rates described in the literature, ranging 10-30% (5). Moreover, epidemiological studies in Iran have shown rates of GBS colonization as 5.3-26.7% (15-23).
The study performed by Fatemi et al. reported rates of GBS colonization as 19.7% by PCR using 16S rRNA and 20.6% by culture method. Sensitivity and specificity were reported 82.3% and 96.5%, respectively (21). Another recent study performed by Bakhtiari et al. resulted a 9.3% prevalence of GBS in culture method and 11.2% by PCR, using the cfb gene. Both sensitivity and NPV were 100% and specificity and PPV of PCR were 98% and 100%, respectively (24). According to the study by Fabien Rallu et al. the prevalence of GBS was determined 16% by culture (sensitivity 42.3%, specificity 100%, PPV 100%, NPV 74.3%), 28% by PCR using thecfb gene (sensitivity 75.3%, specificity 100%, PPV 100%, NPV 87.1%), 37% by PCR using the scpB gene (sensitivity 99.6%, specificity 100%, PPV 100%, NPV 99.7%), 22% by GBS antigen detection (sensitivity 57.3%, specificity 99.5%, PPV 98.5%, NPV 79.5%) (25). Another study performed by de-Paris et al. showed the prevalence of GBS 26.99% by PCR using theatr gene and 15.96% by culture method. Sensitivity and specificity for PCR were 100% and 86.88%, respectively. NPV was 100% and PPV was 59% (12).
The different prevalence rates between several studies may be associated with gestational age at culturing, differences in sampling sites (i.e. only vaginal vs. rectovaginal), using different culture techniques, using PCR with different targets, an alteration of prevalence with time, or real differences of prevalence in various populations or ethnic groups (26). In addition, differences in the elapsed time after sampling for transporting clinical specimens to be processed, using different nucleic acid extraction method, as well as using PCR with different targeting can be some reasons for differences related to reported specificity and sensitivity of PCR in several studies.
In 27 specimens, the PCR results were positive and the cultures were negative (Table 2). Additional analysis including repeating the assay and using PCR targeting the atr, cfb, and scpB genes demonstrated PCR products consistent with GBS, substantiating that these 27 individuals were true GBS carriers with negative GBS cultures. Thereby, these specimens were considered to be true GBS carriers in whom culture method failed to detect GBS.
Interpretation of PCR results associated with cultures is challenging when a positive result for bacterial nucleic acids is compared with negative result for culture. The reason for this contrary may be related to detection of bacteria as a result of a higher sensitivity of PCR for detection of circulating nucleic acids, or non-proliferating, dead or degraded GBS (viable versus nonviable GBS); for instance, in antibiotic pretreated individuals. Other possibilities regarding this subject may be associated with remaining nucleic acid (microbial DNA aemia) mainly after successful antimicrobial therapy; in fact, the remaining nucleic acid may be detectable up to several days without evident clinical significance or positive culture result (12, 27, 28). Besides, presenting some none-GBS bacteria in the vaginogenital tract, such as enterococci or other streptococci species can inhibit the growth of GBS even when using a selective medium such as Lim broth. In addition, individuals may have a very low bacterial load during labor which leads to a small number of GBS cells in the swab samples. In addition, collection, storage and transportation errors are other possible considerations which might affect the culture growth particularly in individuals with a small number of GBS cells. On the other hand, 5% of all GBS strains are nonhemolytic, and in such cases, the GBS colony/colonies may hide from visualization or differentiation in the burden colony of other presenting nonhemolytic streptococci or enterococci colonies. The final possible explanation for these differences is the fact that using double-sampling for each detection method separately may have influenced the intrapartum culture and PCR as the load of bacteria might have been different in the two sampling swabs, leading to discrepancy in yielded results of the two methods.
Another discrepancy about this study was the results of the two samples, for which culture was positive and PCR was negative. PCR was repeated and also tested using the three other mentioned genes to confirm the true yielded results. Possible explanation for these results might be the failures association with the method, such as presence of inhibitors for PCR reaction and probable inaccuracy in handling, collection and transportation (6). In addition, more detailed reviewing of culture plates showed that the two plates grew one colony; thereby this discrepancy can be associated with the lower limit of detection (sensitivity) of PCR. Therefore, these were considered false-negative results.
It is required to select species-specific pairs of primers to apply accurate PCR diagnosis as well as minimize false positive results in direct clinical specimens. An advantage of using PCR targeting 16S rRNA as a gene for detecting bacteria is the high concentration of the 16S rRNA gene targets in bacterial cells. In fact, the presence of multiple copies of genes encoding rRNA in the genome of a given bacterium allows for more sensitive detection through the multiple target sites. However, due to high homology of the 16S rRNA gene regions in different species as well as the presence of several copies of this gene (intragenomic copies) that can differ in sequence, it may lead to identification of bacteria other than GBS and thereby ending up with false positive results (27, 29), as shown in this study.
In conclusion, this study demonstrated that performing only culture method for detecting GBS in pregnant women leads to missed false negative carrier individuals' detection. Therefore, it is recommended that both PCR and conventional culture method be routinely performed to detect GBS in pregnant women accurately. Furthermore, PCR diagnosis demonstrated more sensitive results and shorter turnaround time compared with culture method for detection of GBS colonization in pregnant women. In fact, PCR has the potential for direct intrapartum detection of GBS colonization.