1. Background
Acinetobacter spp. are obligate aerobic, oxidase-negative, non-motile, Gram-negative coccobacilli and opportunistic pathogens that can be easily isolated from soil and water, and sometimes from the hospital environments (1, 2). They don’t require a specific nutrient medium to survive. Acinetobacter spp. grow easily in conventional laboratory mediums without any pigment (3, 4). Due to its low nutritional requirements for growth, this bacterium can survive for long periods in adverse conditions, dry surfaces as well as in aquatic environments (5). Acinetobacter baumannii is one of the most common pathogenic bacteria causing nosocomial infections in patients hospitalized in intensive care units. People with neutropenia, cystic fibrosis, and immune deficiency are exposed to the risk of infection with A. baumannii (6). The catheter and other medical equipment may lead to the outbreak of this bacterium in the hospital (7). Owing to the high levels of antibiotic resistance in comparison with other nosocomial isolates and its high prevalence in hospital environments, A. baumannii is known as an important cause of disease, mortality and economic loss in different countries (8, 9). Besides respiratory tract infections, A. baumannii may also responsible for urinary tract and wound infections in hospitals (10). The resistance of A. baumannii strains to antibiotics can be intrinsic or through the obtaining of genetic factors. Most of them are resistant to ampicillin, amoxicillin/clavulanic acid, antistaphylococcal penicillin, extended-spectrum cephalosporins (except ceftazidime and cefepime), tetracycline, macrolides, rifampicin and chloramphenicol (11, 12). As reported in most hospitals worldwide, multidrug-resistant (MDR) A. baumannii has recently become a major concern in hospitals (13). The prevalence of MDR A. baumannii to other sites indicates the role of this organism in the rapid spread of resistance genes (14-16). Beta-lactamases are inactivating enzymes for the beta-lactam antibiotics. The first identified beta-lactamase is penicillinase (17-21). CTX-M type beta-lactamases, firstly identified in Germany in 1989, consist of a group of Extended-spectrum β-lactamases (ESBLs) encoded by the plasmid (22). Based on the amino acid sequences, CTX-M beta-lactamases are classified into five major groups: CTX-M1, CTX-M2, CTX-M8, CTX-M9, and CTX-M25 (23). The CTX-M enzyme mainly hydrolyzes cefotaxime and often has poor activity against ceftazidime. However, CTX-M15 has a strong activity against ceftazidime (24). The prevalence of ESBLs, especially CTX-M, has increased in recent years (25). These enzymes result in the resistance of bacteria to penicillin and a wide range of third-generation cephalosporins. However, ESBLs are sensitive to several antibiotics such as cephamycin and carbapenem. Also, some of the antibiotics such as clavulanic acid, tazobactam and sulbactam completely inhibit these enzymes (26). Acinetobacter baumannii is inherently capable of producing class D oxacillins (belonging to the OXA-51 group of enzymes) and non-induced AmpC cephaloporinases (27). The blaPER-1 gene was first found in Pseudomonas aeruginosa. It has since been widely observed in Acinetobacter (28). The blaVEB-1 gene was first observed during the outbreak of clonal strains of A. baumannii in the ICU of a French hospital (29). Pumping of the drug out of the bacteria by efflux mechanisms is also related to the specificity of MDR. The tetA is involved in resistance to tetracycline and tetB is involved in the minocycline pump in addition to tetracycline (27, 30). VIM-type beta-lactamase in Acinetobacter species was first observed in Europe and then reported worldwide (31, 32). NDM beta-lactamase is a transmissible class B molecular β-lactamase recently identified in New Delhi, India (33). Beta-lactam resistance genes in A. baumannii are usually located on mobile genetic elements and therefore can easily transmit between different strains. Thus, identification of the ESBLs-producing strains can be an essential step in the treatment of their infections.
2. Objectives
Due to the high prevalence of antibiotic resistance genes and nosocomial infections, the aim of the present study was to isolate A. baumannii from clinical specimens and determine the prevalence rate of blaOXA-51, blaNDM, blaVIM, blaPER, blaVEB, blaCTX-M, tetA and tetB genes in the isolates.
3. Methods
This study was descriptive cross-sectional research performed on 129 clinical isolates of Acinetobacter isolated from blood, urine, wound exudates and respiratory secretions in the hospitals in Tabriz city from March to December 2019. All of the isolates were detected using standard laboratory tests, microbiological experiments and differential biochemical tests including oxidase, catalase, urease, oxidation-fermentation (OF), Triple Sugar Iron (TSI) tests, culture in Simmons citrate agar, sulfur indole motility media (SIM), Methyl red-Voges-Proskauer (MR-VP) broth as well as, the growth in 37°C and 42°C. Antibiotic resistance patterns of A. baumannii isolates were determined by agar disc diffusion method based on the guidelines of the clinical & laboratory standards institute (CLSI) (34). The used antibiotic disks are included: Ceftazidime (30 μg), ciprofloxacin (5 μg), levofloxacin (10 μg), cefotaxime (30 μg), Aztreonam (30 μg), cefepime (30 μg), gentamicin (10 μg), amikacin (30 μg), imipenem (10 μg), meropenem (10 μg), piperacillin/tazobactam (10/100 μg), tetracycline (30 μg) ) and polymyxin B (300 μg) (Mast Diagnostics Mast group Ltd., Merseyside, UK). The standard strain of A. baumannii ATCC 19606 and Escherichia coli ATCC 25922 was used as positive and negative controls, respectively. The ESBL-producing isolates were identified by combined disk test using ceftazidime (30 μg), cefotaxime (30 μg), ceftazidime/clavulanic acid (30 μg/10 μg) and cefotaxime/clavulanic acid (30µg/10µg). After incubation at 37°C for 24 hours, ESBL-producing isolates had a zone of inhibition with a diameter ≥ of 5 mm around ceftazidime/clavulanic acid discs in comparison with ceftazidime or cefotaxime discs (35). The DNA samples were extracted from the isolates using a commercial kit (Invitek, STRATEC Molecular-Germany). To identify the blaOXA-51, blaNDM, blaVIM, blaPER, blaVEB, blaCTX-M, tetA and tetB genes in the isolates, the polymerase chain reaction (PCR) technique was performed using the specific primers (Table 1). The reaction was performed in a total volume of 20 µL, including 10 µL of Master Mix (Amplicon Denmark), 2 µL of template DNA, 10 pM of primer and distilled water. The heating program in thermocycler was as follows: One cycle of initial denaturation at 94°C for 3 min, 35 cycles of denaturation in 35°C for 30 sec, annealing at 45°C for 1 minute, and extension at 72°C for 1 min. The final extension was carried out at 72°C for 4 minutes. The PCR products were run on 1% agarose gel in TBE buffer for 60 min at 100°C. The gel was then placed in a tank containing ethidium bromide for 15 minutes. The results were visualized using a gel documentation system under the UV light. Standard strains of A. baumannii ATCC 19606, P. aeruginosa ATCC 27853 and E. coli ATCC 25922 were used as quality control according to the CLSI.
| Genes | Sequences | Amplicon Size | References |
|---|---|---|---|
| blaOXA-51 | 5′-TAA TGC TTT GAT CGG CCT TG-3′ | 353bp | (36) |
| 5′-TGG ATT GCA CTT CAT CTT GG-3′ | |||
| blaNDM | 5'-GGTTTGGCGATCTGGTTTTC-3' | 621bp | (37) |
| 5'-CGGAATGGCTCATCACGATC-3' | |||
| blaVIM | 5'-ATTGGTCTATTTGACCGCGTC-3' | 514bp | (38) |
| 5'-AATGCGCAGCACCAGGATAG-3' | |||
| blaPER | 5'-GTTAATTTGGGCTTAGGGCAG-3' | 855bp | (39) |
| 5'-CAGCGCAATCCCCACTGT-3' | |||
| blaVEB | 5'-CGACTTCCATTTCCCGATGC-3' | 643bp | (40) |
| 5'-GGACTCTGCAACAAATACGC-3' | |||
| blaCTX-M | 5'-CGCTTTGCGATGTGCAG-3' | 550bp | (41) |
| 5'-ACCGCGATATCGTTGGT-3' | |||
| tetA | 5'-GCGCGATCTGGTTCACTCG-3' | 164 bp | (42) |
| 5'-AGTCGACAGYRGCGCCGGC-3' | |||
| tetB | 5'-TACGTGAATTTATTGCTTCGG-3' | 206 bp | (42) |
| 5'-ATACAGCATCCAAAGCGCAC-3' |
Sequence of Primers Used
4. Results
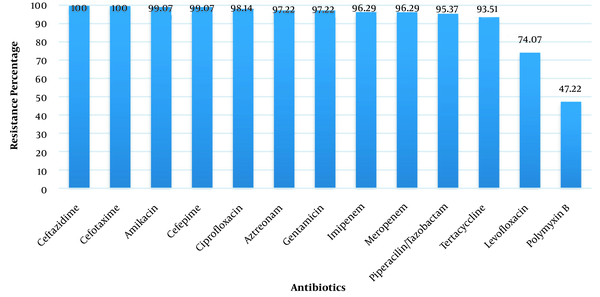
Out of 129 isolates, 108 (83.72%) isolates were identified as A. baumannii. The mean age of patients with A. baumannii infections was 52 ± 24.4 years. The organism was isolated from 58 (53.7%) of urine cultures, 10 (9.26%) of wound discharges, 34 (31.48%) of blood cultures and 6 (5.56%) of respiratory tract secretions. The isolates showed the least resistance to polymyxin B (47.22%). The highest resistance of them was detected to cefotaxime (100%) and ceftazidime (100%) (Figure 1). Out of 108 isolates, 103 isolates (95.37%) were identified as MDR (resistant to more than three classes of antibiotics). The results of the combined disk test showed that 14 (12.96%) isolates were ESBL-positive. PCR results for target genes showed that 43 (39.81%) isolates contained tetA gene, 35 (32.40%) isolates contained blaPER-1 gene, 34 (31.48%) isolates contained blaCTX-M gene, 23 (21.29%) isolates contained tetB gene, 20 (18.51%) isolates contained blaNDM gene,18 (16.66%) isolates contained blaVEB-1 gene and 18 (16.66%) isolates contained blaVIM gene. The blaOXA-51 gene was positive as a genetic marker for the diagnosis of A. baumannii in all isolates. The ESBL-producing isolates showed the highest resistance to the used antibiotic discs among others.
5. Discussion
Production of beta-lactamases by gram-negative bacteria is one of the main mechanisms responsible for their resistance to beta-lactam antibiotics. Since the beta-lactams have wide clinical applications, beta-lactamases have evolved concurrently and played a major role in the failure of antibiotic therapy (43). In the last fifteen years, epidemics of infection caused by beta-lactamase-producing organisms have occurred around the world. So, these enzymes are known as a major threat to the use of cephalosporins. It has also been well established that the treatment of such cephalosporin-resistant infections will not be satisfactory, and the mortality caused by ESBL-producing bacteria is significantly high (44). The emergence and prevalence of ESBL-producing bacteria appear to be due to the widespread use of extended-spectrum beta-lactams. The prevalence of these bacteria in different parts of the hospital has been increased in recent years. In the present study, out of the 129 Acinetobacter isolates, 108 (83.72%) were identified as A. baumannii. This result is almost similar to the findings of Nazari Monazam et al. (45) (76.9%), Constantiniu et al. (46) (71%) and Rit and Saha (47) (74.02%). However, Ahmadikia et al. (48) reported higher rates (93.1%) than of the present study. In this study, the antimicrobial resistance analysis indicated that all isolates were resistant to ceftazidime and cefotaxime. Ayan et al. (49) found that all 52 isolates were resistant to piperacillin/tazobactam, cefepime, cefotaxime, ceftazidime, gentamicin, and aztreonam. The resistance to aminofloxacin and tetracycline were reported in 8% and 74% of strains, respectively. Biendo et al., in a study about the antibiotyping of A. baumannii isolates, found that 15 of 18 isolates were resistant to ticarcillin, ticarasilamine/clavulanic acid, piperacillin/tazobactam, ceftazidime and aztreonam (50). Their findings are quite consistent with the results of the present study. In the study of Smolyakov et al., (51) and Wang et al., (52), all strains were resistant to imipenem. Also, Saadatian (2005) reported that 95.5% of A. baumannii isolates were resistant to amikacin. These results were is consistent with the findings of the present study (53). In the present study, the resistance level of A. baumannii isolates to polymyxin B was high, so that 47.22% of isolates were resistant to this antibiotic. Polymyxins are the last-line treatment for MDR isolates of A. baumannii. Therefore, treatment of A. baumannii infections, which is resistant to these antibiotics, is very difficult (54). In the present study, 103 isolates (95.37%) had MDR that was higher than the findings of Joshi (55) (75%) and Bahador (56) (45%) and less than Ahmadikiya (98.9%) (48). In this study, the prevalence of ESBL-producing A. baumannii in clinical specimens was 12.96%, which is higher than the results of Ahmadikiya (48) and lower than the findings of Sinha (57) in India and Maleki (58) in Shiraz. Ranjbar and Farahani in their study of 163 strains of A. baumannii showed that 52.2% of the samples were ESBL positive. Which is more than the findings of the present study (59). In the study of Ahmadikiya et al. (48), 31.6% of isolates were positive for the blaCTX-M gene, which is similar to the findings of present study. Also, in the study of Shahcheraghi et al. (60) and Celenza et al. (61), these rates were 1.2% and 30.4%, respectively, which are lower than the findings of the present study. In the present study, the presence of the blaOXA-51, blaNDM, blaVIM, blaPER, blaVEB, blaCTX-M, tetA and tetB genes are 100%, 18.51%, 16.66%, 32.40%, 16.66%, 31.48%, 32.40% and 21.29%, respectively. Safari et al reported that 58% and 20% of ESBL-positive A. baumannii isolates contained SHV and CTX-M genes, respectively (62). Goudarzi et al. In 2016, the resistance of isolated strains to tested antibiotics was 95.4% to ceftazidime, 100% to cefotaxime, 95.7% to cefepime, 91.7%to imipenem, 91.7% to meropenem, 80.6% to amikacin, 97.2%to piperacillin, 92.6%to ciprofloxacin, 95.4% to piperacillin/tazobactam, 40.7% to gentamicin, 98.1% to ampicillin/sulbactam and 98.1% to co-trimoxazole respectively. PCR results showed that 44.17% of the isolates had blaVIM gene and blaNDM gene was not seen in the strains (63). Mohammadi et al. Showed in a study that antibiotic resistance in 100 isolates of A. baumannii was related to antibiotics: Cefimoimide (97%), Ceftriaxone (95%), Amikacin (95%), Imipenem (76%), Piperacillin-tazobactam (70%), Meropenem (69%), Gentamicin (63%), Tobramycin (56%), Tetracycline (51%), and Ampicillin-Sulbactam (49%) and lowest resistance was related to polymyxin B. PCR results showed that 17% and 20% of the strains carried blaVIM and blaNDM genes, respectively (64). Azizi and Shahcheraghi during a study in 2017 in Tehran hospitals, showed that all samples were resistant to gentamicin, ciprofloxacin, piperacillin, cefotaxime, ceftazidime and tetracycline. Also, all isolates were identified as resistant to several antibiotics. The tetA, tetB, blaVEB, blaCTX-M and blaPER genes were identified as 75.3%, 43%, 35.3%, 76.9% and 61.5% of the isolates, respectively (65). In a 2012 study by Asadollahi et al., The prevalence of tetA and tetB genes was reported to be 95.5% and 65%, respectively (66). The blaOXA-51 gene is located on the chromosome in A. baumannii. The enzyme OXA-51 has poor carbapenemase activity, but the addition of the ISAba 1 complement sequence at the 5 'end of the blaOXA-51 gene leads to its high expression, and this increase in expression causes resistance to carbapenem (67). In 2015, Badmasti et al reported a 44% presence of the blaPER gene in A. baumannii isolates. Which is more than the findings of the present study (68). Ranjbar and Farahani during a study in 2019, the prevalence of blaOXA−23, blaVIM, blaPER−1 and tetB genes was reported to be 85.1%, 60.5%, 42.3% and 67.8%, respectively (59). The differences between the findings of the present study and other researchers may be due to differences in the place of sample collection, the number of samples studied and even the decrease or increase in antibiotic use in the patients studied
5.1. Conclusions
According to the results of this study, the resistance of A. baumannii isolates to various antibiotics especially beta-lactams is an important therapeutic problem. Also, the production of ESBLs is a major threat for use of extended-spectrum cephalosporins. Therefore, in order to treat infections that are suspected for ESBL production, the appropriate antibiotic should be selected based on the results of the antibiogram test. The findings of the present study indicate the need for making the right decision about the reasonable administration of drugs as well as using novel diagnostic methods in microbiology laboratories.