1. Background
Gram-positive Pseudomonas aeruginosa, a motile bacterium, is well known as a nosocomial pathogen able to infect almost all tissues. It is an aerobic bacillus that belongs to the Pseudomonadaceae family, which can cause acute as well as chronic infectious causing 18 to 63% of infections and diseases worldwide (1, 2). Pseudomonas aeruginosa can defend against diverse physical conditions, and thus can survive in community and hospital backgrounds. It is a cause of various infections i.e. urinary tract infections (UTIs), pneumonia and surgical site infections. It is considered a primary contributing factor to lung infection in cystic fibrosis patients. It can produce biofilm and stick to the lower respiratory tract of human mucin (3). Pseudomonas aeruginosa can be eliminated only at the beginning of colonization whereas, a preferable decrease in bacterial density during the colonization to prevent exacerbations (4).
Furthermore, P. aeruginosa is responsible for community-acquired infections including ulcerative keratitis, otitis externa and soft tissue infections (5). It is a main health danger, particularly to immune-deficient patients (6). This organism can simply acclimatize to the variations in the environment, hastily build up antibiotic resistance, and generate a range of virulence factors and thus it is connected with significant morbidity. Pseudomonas aeruginosa holds a range of virulence genes to assist in disease progression and colonization among a diversity of environments. Pseudomonas aeruginosa is a highly resistant pathogen due to its limited susceptibility to certain antimicrobials, which is one of its main pathogenic characteristics. In fact, this is reflected that the majority of resistance genes that are currently known are present in the genome of P. aeruginosa (7, 8).
The genes nan1, toxA, exoS, exoU, oprL, plcN, oprI, lasA, exoY, lasB, plcH, and oprD are typically responsible for and contribute to the virulence factors (9). OprL as well as OprI are outer membrane proteins (OMPs) of P. aeruginosa which play a significant role in the interaction of the bacterium with its environment and also contribute to the innate resistance of P. aeruginosa to antibiotics.
Additionally, the occurrence of particular OMPs is concerned in the transport systems i.e. efflux which contributes to cell permeability (10, 11). According to a report, OprL has a major impact on the creation of membrane vesicles in P. aeruginosa and is a reliable indicator of P. aeruginosa infection (12). This second most abundantly present OMP of the P. aeruginosa aids in maintaining the bacterial cells’ integrity and is also a part of the Tol-OprL multiprotein system where it plays an important role in the transport of carbon sources across cytoplasmic membranes (13-15). Similarly, P. aeruginosa has a lasB gene that produces Elastase B, a multifunctional protease which is involved in degrading host defence, immunoregulatory proteins as well as epithelial damage and type III secretion system (16). lasB gene is considered to be a prospective target for the advancement of a novel chemotherapeutic approach, particularly against multidrug-resistant strains (11).
In the veterinary field, antibiotics are administered abundantly for curative as well as prophylaxis purposes (17). Careless use of disinfectants also leads to bacterial adaptations and promotes the proliferation of highly resistant bacteria. Intrinsic resistance of P. aeruginosa has led the community as well as nosocomial infections caused by this pathogen, which is a serious concern. In addition to this, P. aeruginosa can acquire new resistance mechanisms against a variety of groups of antibiotics like, β-lactam antibiotics, fluoroquinolones and aminoglycosides (18). Hence, it is crucial to enhance our understanding of the resistance patterns and virulence of particular strains of P. aeruginosa (19). Thus, to mitigate the spreading risk of the resistant P. aeruginosa strain, it is crucial to obtain antimicrobial susceptibility patterns of the P. aeruginosa isolates (20). Moreover, the speedy identification of the strains of bacteria responsible for hospital-acquired infections is particularly crucial for making informed treatment decisions for patients. Also, animals and foodstuffs from animals are involved in daily human lives, increasing the risk of transmission of pathogenic strains between humans and animals.
2. Objectives
This study aims to identify presence of two virulence genes, OprL and LasB, in P. aeruginosa strains isolated from the samples obtained from the hospital and veterinary laboratories.
3. Methods
3.1. Isolation of Bacteria and Biochemical Test
One hundred twenty specimens of P. aeruginosa were collected from various hospitals and veterinary laboratories in Ardabil, Iran. Out of these 60 samples were from the hospital and the rest of the 60 samples were collected from the veterinary laboratories. Human clinical samples include; exoda of pulmonary, sputum, blood and abscess and veterinary samples were milk samples.
3.2. Sample Processing
The collected samples were cultured on Nutrient and MacConkey agar media and then incubated at 37°C for 24 h to ensure purity and optimal growth. The isolation procedure for P. aeruginosa was conducted using the Shukla and Mishra protocol from 2015 (21). Colonies that showed suspicious growth and colony morphology were identified. Additionally, conventional biochemical tests such as catalase test, motility, oxidase test, indole sulfide (SIM), and triple sugar agar (TSI) were performed to identify P. aeruginosa (22).
3.3. Bacterial DNA Preparation
Pseudomonas aeruginosa DNA was extracted using a commercially available kit, Cinnapure DNA (Cinnagen, Tehran, Iran), according to the instructions labelled by the manufacturers.
3.4. Detection of Virulence Genes by Polymerase Chain Reaction
The prevalence of virulence genes oprL, and lasB was determined by polymerase chain reaction (PCR) (23). Specific primers listed in Table 1 are used to amplify virulence genes. As per instructions provided by the supplier, primers were in a lyophilised form dissolved in the nuclease-free water to make up the final concentrations up to 100 pmol/mL. A 25 µL reaction mixture was prepared to conduct PCR amplification of the oprL and lasB genes, which included 0.5 µL of dNTPs (10 mm), 0.5 µL of each primer (10 pmol), 1.5 µL of MgCl2 (25 mm), and 0.2 µL of Taq DNA polymerase (5 U/µL). For PCR amplification of oprL and lasB genes, a 25 µL reaction mixture was prepared containing 0.5 µL of dNTPs (10 mm), 0.5 µL of each primer (10 pmol), 1.5 µL of MgCl2 (25 mm), and 0.2 µL of Taq DNA polymerase (5 U/µL). Both the genes were amplified separately. Strain of P. aeruginosa ATCC 27853, was used as a positive control. Agarose gel electrophoresis with a 1% agarose gel stained with ethidium bromide was utilized to visualize the PCR products.
| Primers | PCR Program | Primer Nucleotide Sequence | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| oprL | 30 cycles: 40 s 94°C, 40 s 57°C, 50 s 72°C | F: 5’-ATGGAAATGCTGAAATTCGGC-3’ | 504 | Xu et al., 2004 (24) |
| R: 5’-CTTCTTCAGCTCGACGCGACG-3’ | ||||
| lasB | 35 cycles: 30 s 94°C, 40 s 57°C, 50 s 72°C | F: 5’-TTCTACCCGAAGGACTGATAC -3’ | 153 | Zhu et al., 2004 (25) |
| R: 5’-AACACCCATGATCGCAAC-3’ |
4. Results
4.1. Isolation and Detection of Pseudomonas aeruginosa
About 120 specimens P. aeruginosa clinical isolates were collected and the identification of these samples was confirmed using conventional bacteriological techniques. Results of the bacteriological test from 76 samples exhibit the green and blue colonies with the sweet grape like scent, indicating the presence of Pseudomonas produced on MacConkey agar and it didn’t ferment lactose sugar. Pseudomonas aeruginosa strains were present in 24 (40%) animal sample isolates and 52 (86%) human sample isolates (Table 2).
| Sl. No. | Specimens | Number of P. aeruginosa Isolates; No. (%) |
|---|---|---|
| 1 | Animal samples (60) | 24 (40) |
| 2 | Human samples (60) | 52 (86) |
4.2. Molecular Detection of the Virulence Genes
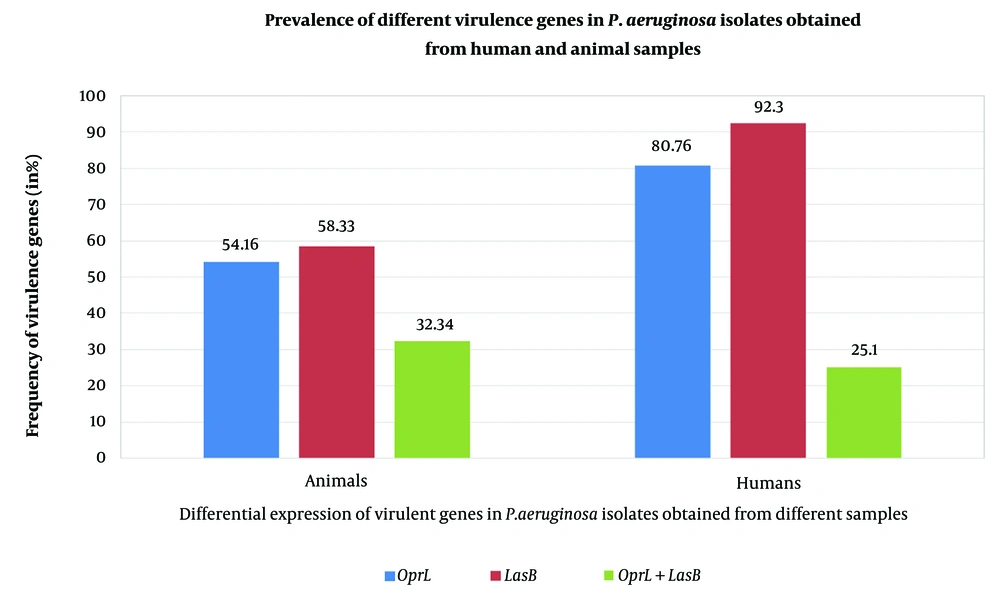
Polymerase chain reaction analysis done to screen 2 virulence encoding genes, lasB and oprL. The testing was conducted only for P. aeruginosa isolates, and specific primers were utilized for targeting virulence gene i.e. oprL and lasB that results in the amplicons of 504bp and 153 bp. The oprL genes were detected in 54.16% of animal sample isolations and 80.7% of human sample isolates. Results highlighted that 58.33 % of animal sample isolates and 92.30 % of human sample isolates showed the presence of lasB genes. However, PCR results showed the coexistence of both the virulence genes in 32.34% of animal sample isolates and 25.10% of human sample isolates (Figure 1).
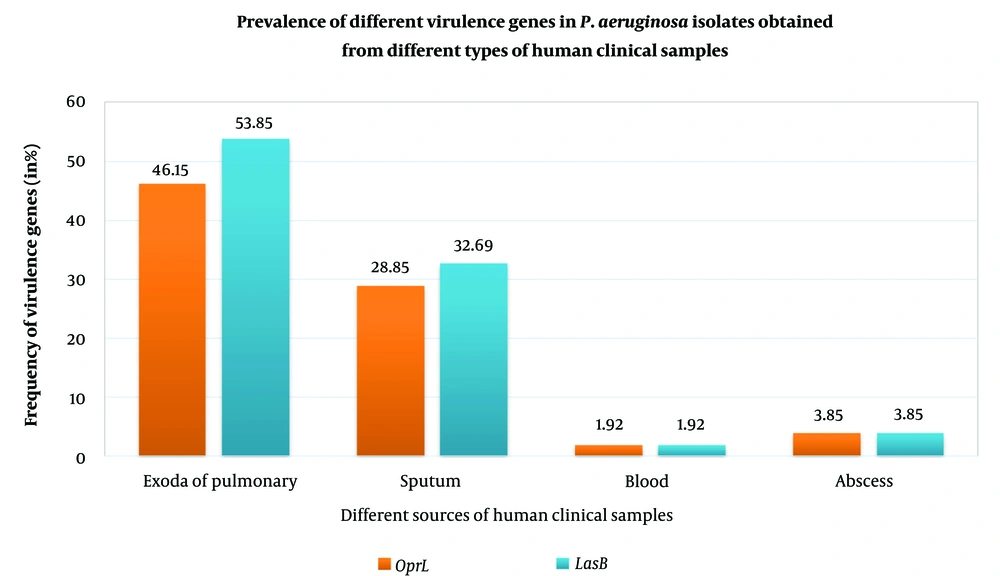
The expression of oprL and lasB were higher in the exoda of pulmonary compared to all other samples and the least expression was found in abscess and blood. Isolates from sputum showed the prevalence of the genes oprL and lasB, by more than 50% (Figure 2).
4.3. Statistical Analysis
We have used a descriptive statistics for the analysis, to know the prevalence of virulence genes; lasB and oprL, in all of P. aeruginosa isolates obtained from both human and animal clinical samples collected from the hospital and the veterinary laboratories. With the help of comparative analysis, the prevalence of these genes in the different isolates obtained from different clinical samples, were also investigated.
5. Discussion
The ubiquitous and dangerous nature of P. aeruginosa can be attributed to genetic adaptability, its ability to acquire antimicrobial resistance AMR, and its capacity to express various virulence factors. Additionally, its capacity to form biofilms adds to its notoriety as a pathogen (26). Numerous studies reported the disease outbreaks caused by P. aeruginosa that accredited both the environmental source as well as hospital-acquired infections. Pseudomonas aeruginosa is found in environmental sources and is linked to severe infections in immunocompromised hosts, patients with severe burns, and those with surgical injuries (27, 28). This situation is due to the prospective colonization factors such as pilli and the alginates and extensive drug resistance strains of P. aeruginosa in the hospitals (29). Moreover, this organism is one of the most vital sources of disease in patients suffering from cystic fibrosis. Estimates reveal that more than 80% of cystic fibrosis patients are infected with P. Aeruginosa (30). Pseudomonas aeruginosa can consume a wide range of organic materials as energy sources and hence it can exist in the environment over an extended period (31). So the exact quick detection of P. aeruginosa and the information about its susceptibility profile is of great importance.
Pseudomonas aeruginosa draws interest as a dreadful pathogen for consumer health of a variety of diseases in humans as well as from the food obtained from animals and is a source of multi-drug resistant characters that are interchangeable in pathogens of humans and animals. One of the major struggles tackling the world is resistance to antibiotics. Hence it is vital to identify P. aeruginosa specifically and rapidly and also recognize the susceptibility patterns of P. aeruginosa strains. This may evade unnecessary treatments with less effective antibiotic and prevents the development of pathogens that are resistant to antibiotics (32). Pathogens use the fabrication of virulence factors during early colonization and acute infection as a survival strategy to evade the host's immune system. This is an important mechanism that allows the pathogen to persist in the host. A large number of the virulence traits and the cell-associated or secreted compounds of both low and high molecular weights are identified as crucial in initiating infections caused by P. aeruginosa (33, 34). While such virulence traits and cell-associated or secreted compounds play a significant role in promoting growth and persistence, they can also cause severe damage to host tissues and weaken immune responses (35). Earlier studies used as a basis to screen various virulence genes in the P. aeruginosa isolates obtained from the different samples. Billard-Pomare and coworkers' study showed the real-time PCR of the oprL gene has a higher specificity and is a more suitable one than the culture for demonstrating bacterial colonization (36). Similarly, another study compared the efficacy of PCR and traditional culture for colonization determination and concluded that PCR has elevated specificity as well as sensitivity in comparison to the culture (37). In 2014, another study proved that some sputum samples tested positive by the PCR polymerase chain reaction analysis, while their cultures were found negative. Polymerase chain reaction samples become positive prior in comparison with the culture examinations in most of the cases as the samples transformed to positive at some stage in the period of hospitalization. Thus, they suggested that the PCR method has adequate sensitivity as well as specificity to distinguish colonized patients (38).
Of the virulence genes studied in this study, the oprL gene along with maintenance of bacterial cell integrity recently has been found to play a significant role in protecting bacterial cells from oxidative stress and also play a crucial role in conferring resistance against different antiseptics and antibiotics (13-15, 24). Also, molecular identification of the oprL gene with PCR provides a rapid and reliable method to identify P. aeruginosa strains (39). Another virulence gene studied in Pseudomonas aeruginosa; lasB encodes for the most important protease an extracellular collagenase (an elastase enzyme). It contributes to the bacterial virulence mechanism by cleaving the components like elastin or collagen of the host tissues along with disruption of the cellular junctions to cause host tissue damage aiding bacterial invasion. It can break down immunoglobulins G and A and is also involved in the destruction of fibronectin, which can expose ligands for bacterial attachment (40, 41). Also, some studies showed its role in immune modulation along with another protease via inhibition of flagellin-mediated immune system recognition and detection. The presence of the lasB gene in P. aeruginosa isolates isolated from different environments and clinical samples, indicates its significance in the survival of the bacteria in diverse settings. lasB mutation reduces the invasion of P. aeruginosa and so molecular detection of this gene also helps to identify P. aeruginosa (24, 40, 42, 43).
So in the current study, overall 76 isolates of P. aeruginosa were curtained for the occurrence of the specific virulence genes, oprL and lasB. Among 24 animal sample isolates, the oprL genes were detected in 54.16% and lasB genes in 58.33%. The coexistence of both virulence genes was observed in 32.34% of animal sample isolates. On the other hand, 80.76% of human sample isolates expressed oprL genes and 92.30% showed the presence of lasB genes. 25.10% of human sample isolates showed the coexistence of both oprL and lasB genes. Pathogenicity associated with P. aeruginosa is obviously multifactorial. According to one study, oprL was promised in AMR antibiotic resistance along with bio-film formation directs for numerous troubles regarding the presence of P. aeruginosa infections can complicate the management and thus make them difficult to treat (44). In the current study, several isolates detected showed the presence of lasB gene and results of this study are in accordance with the earlier studies (45). Our study results suggested that all strains isolated from either human or animal samples harboured at least one of the virulence genes being tested.
The frequency of P. aeruginosa and expression of its virulence genes depend upon different factors like; the nature of its environment and the extent to which it contaminates as well as the immune response of the patients and also the strain virulence (46). Some strains exhibit improved adaptation to certain conditions originating in the infectious sites which may come back to the vast geographical and environmental sources due to differences in distribution of virulence genes. The absence of virulence factor genes in the strain is attributed to the fact that P. aeruginosa infection can result in genome reduction, which increases its ability to adhere host (47). Differences in distribution of virulence factor genes within the populations suggest that certain P. aeruginosa strains may be better adapted to specific circumstances present in certain sites of infection (48).
To sum up, the combination of oprL and lasB genes in PCR provides a reliable method for the identification of P. aeruginosa. The presence or absence of different virulence genes among the P. aeruginosa isolates suggests that they are associated with varying levels of inherent virulence and pathogenicity, which may have different impacts on the disease progression and outcome. Moreover, P. aeruginosa strains; isolated from veterinary samples are studied occasionally. Since animals and animal-related food products are part of daily human lives, it potentially increases the risk of transfer of pathogenic P. aeruginosa strains between them. Hence, a significant advantage of our study was the collection of strains obtained from both human and veterinary samples sequentially, enabling us to determine the prevalence of virulence factor genes, oprL and lasB. Thus, data from or study can be useful for the health care workers as well as for the people working in veterinary areas to develop the proper infection control measurements and to set up a surveillance system to minimise the spread of pathogenic P. aeruginosa isolates.