1. Background
Helicobacter pylori is a gram-negative, spiral form bacterium which primarily colonizes human epithelial cells covering gastric and occasionally duodenal or esophageal mucosal membrane (1, 2). Infections are not only associated with gastric or duodenal ulcers, but also show tight correlation with risk factors for gastric and duodenal cancer (3, 4). It seems that humans are the most important or even the sole host for H. pylori. The infection most probably spread directly among humans through oral-oral or oral-fecal routs and even through the use of improperly disinfected endoscopic devices (5, 6). The bacterium colonizes approximately 50% of human population. However, its prevalence differs internationally as it was reported to be 20 - 40% in developed countries while developing countries; whereas, 70 - 90% of populations in developing nations were shown to be positive for this infection (7, 8). Additionally, there seems to be an uneven distribution in different parts of a country (9). The prevalence of this bacterial infection had been in close association with different risk factors including social, economic, and genetic factors (10).
Plenty of efforts have been made to introduce potential biomarkers specific for H. pylori to offer efficient screening methods for detecting positive individuals. Several bacterial proteins have been identified including those participating in transferring this organism to the mucosal membrane, such as flagellin encoded by fleA and flaB genes (11, 12). Of these factors, urease enzyme, due to its intense enzymatic activity, plays an integral role during pathogenesis of this microrganism (13). UreC is an antigenic subunit of urease protein in H. pylori and it was proven to have the greatest antigenicity among other identified proteins, hence providing a platform for many diagnostic methods (14). The ureC gene possesses 1,710 bp encoding for a protein with 569 amino acids; and has a conserved sequence with 95% similarity between different H. pylori isolates (15). Having the potential to be considered as a specific sequence covering almost all H. pylori isolates, it was selected as a target for our polymerase chain reaction (PCR) assays. In order to establish a reliable platform to prevent and control H. pylori infections and to amend reversible risk factors, it is obviously required to decipher the epidemiologic characteristics of the infection in a certain society.
2. Objectives
Our study was designed to determine the prevalence of H. pylori infections in patients with upper gastrointestinal disorders in Qazvin province, Iran, where there is lack of evidences in this context.
3. Methods
3.1. Subjects and Sample Collection
This study included 100 cases suffering from gastrointestinal problems such as abdominal pain, nausea, constipation, gastric burning etc. who referred to gastroenterology ward in Velayat Hospital, Qazvin, Iran. The project was approved by the research Ethic Committee of Qazvin University of Medical Sciences. Prior to be incorporated in our study, all participants were asked to sign a written consent; and a simple questionnaire was provided which included demographic characteristics of the subjects (sex, age, career, clinical symptoms, residency place, history of hospitalization, and history of antibiotic therapies). Following general examination of patients by a specialist, they were referred to endoscopy ward to achieve a primary diagnosis. Gastroscopies were performed using a video-endoscope device (OLYMPUS). Biopsy samples were collected from those with suspected ulcers and transferred to test tubes specific for rapid urease test (RUT). A peace of the biopsy was also maintained at -20°C for DNA extraction.
3.2. Rapid Urease Test
The test was performed immediately after biopsy preparation. Tissue samples containing the mucosal part of antrum of the stomach were placed in urease medium (Bahar-Afshan, Iran) containing urea, and phenol red as a pH indicator. This system detects urease enzyme produced by H. pylori hydrolyzing urea to ammonia. The reaction causes an increase in pH of the medium resulting in a red color in positive cases and yellow color in negative samples.
3.3. DNA Extraction and Primer Design
Biopsy samples were homogenized in test tubes and used for DNA extraction. The DNA samples were isolated using a commercially available kit (Kosar Biotech Company, Tehran, Iran). The DNA samples were quantified using a nanodrop spectrophotometer. To amplify the target sequence (ureC gene), a specific primer pair was designed using Beacon software. To confirm the primer sequence we used primer-BLAST database from NCBI was used. The primer sequence was as follows; forward: 5ˊ-ACGCCCTTTCTTCTCAAGC-3ˊ, and reverse: 5ˊ-GTTCACAAACTTATCCCCAATCG-3ˊ; which should result in a 191bp amplicon.
3.3.1. Polymerase Chain Reaction
The PCR reactions were performed using a PCR Master Mix (Takara, Japan) in an ABI Thermal Cycler device (Applied Biosystems). Briefly, the mixture in reaction tubes contained 10 µ Master Mix, 1 µL of each forward and reverse primer, 140 ng template DNA, and distilled water to a total volume of 20 µL. The thermal program was as follows: The primary denaturation for 5 min at 95°C; followed by 40 cycles of each 95°C (40 seconds), 57°C (30 seconds), and 72°C (30 seconds); and a final extension for 15min at 72°C. The PCR products were then resolved on a 2.5% agarose gel to confirm the size and specificity of the amplicon.
3.3.2. DNA Sequencing of the PCR Products
In order to identify the positive samples, PCR products were assessed regarding the ureC sequence variations. Prior to sequencing, the PCR products were purified using a commercially available purification kit (Roche, Germany). The nucleotide-BLSAT database from NCBI was used as a tool to align the resulted sequences.
3.4. Statistical Analysis
Statistical data interpretations were performed using SPSS software, version 20. The quantitative data were calculated as the mean ± SD. To analyze the relativity of different variables, a student’s t-test was used’; with P-value of less than or equal to 0.05 as the borderline of statistical significance.
4. Results
A total of 100 patients complaining from upper gastrointestinal symptoms were included and analyzed regarding the H. pylori infection. The subjects were 47% men and 53% women. The age range of the participants was 25 - 60 years for the men and women.
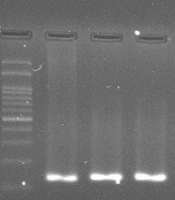
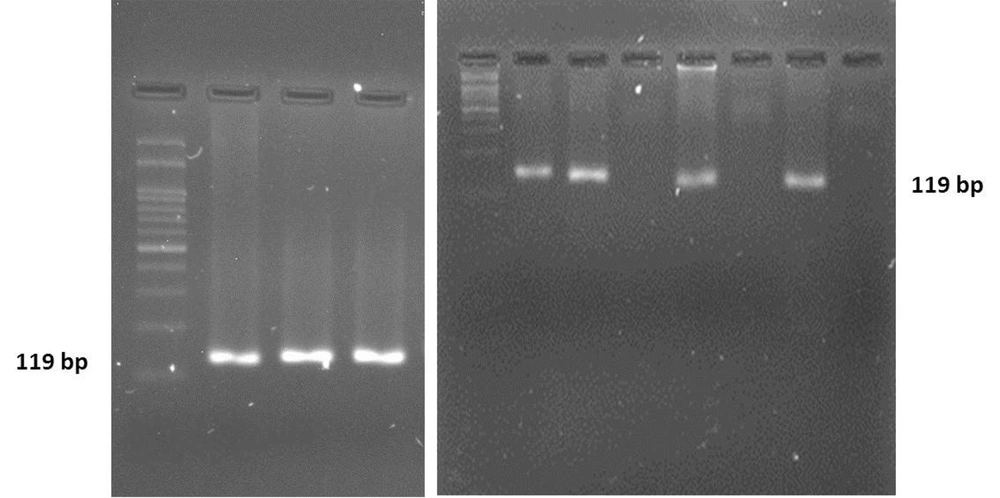
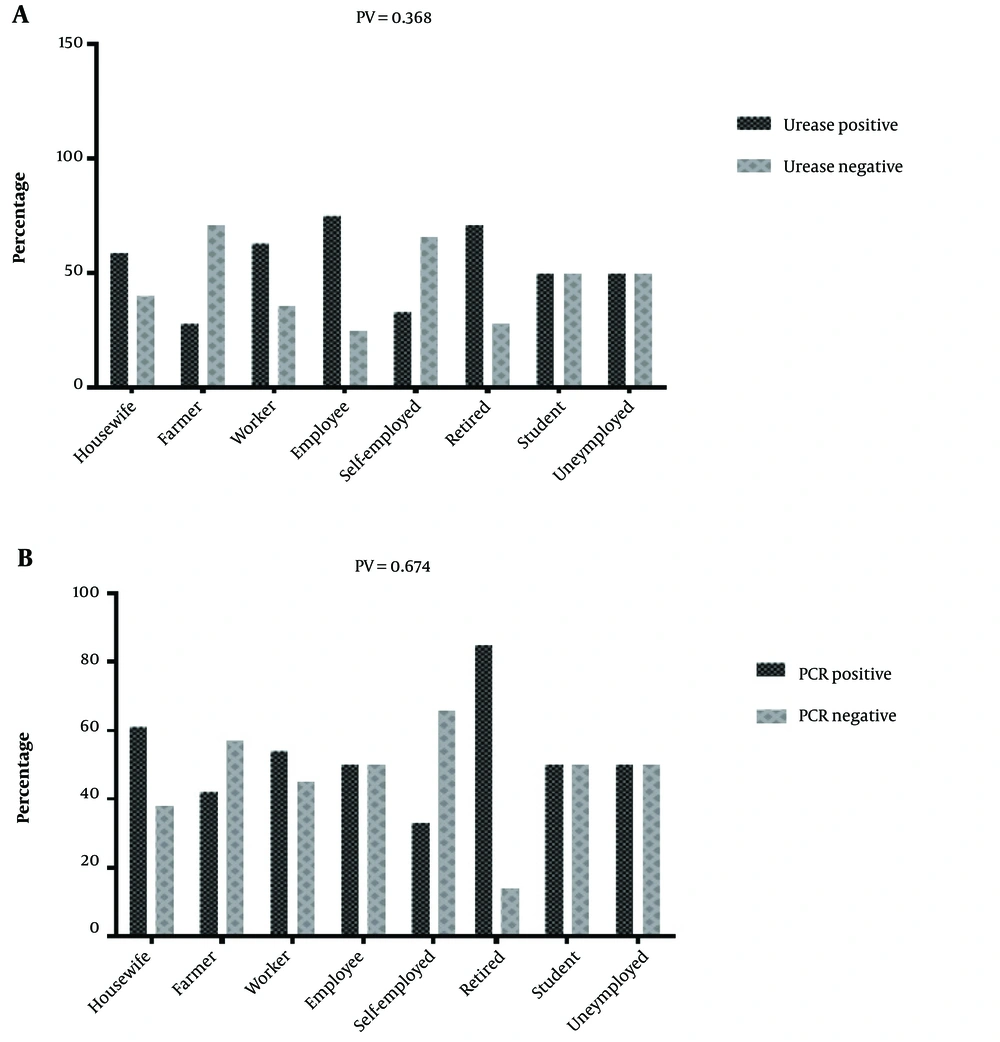
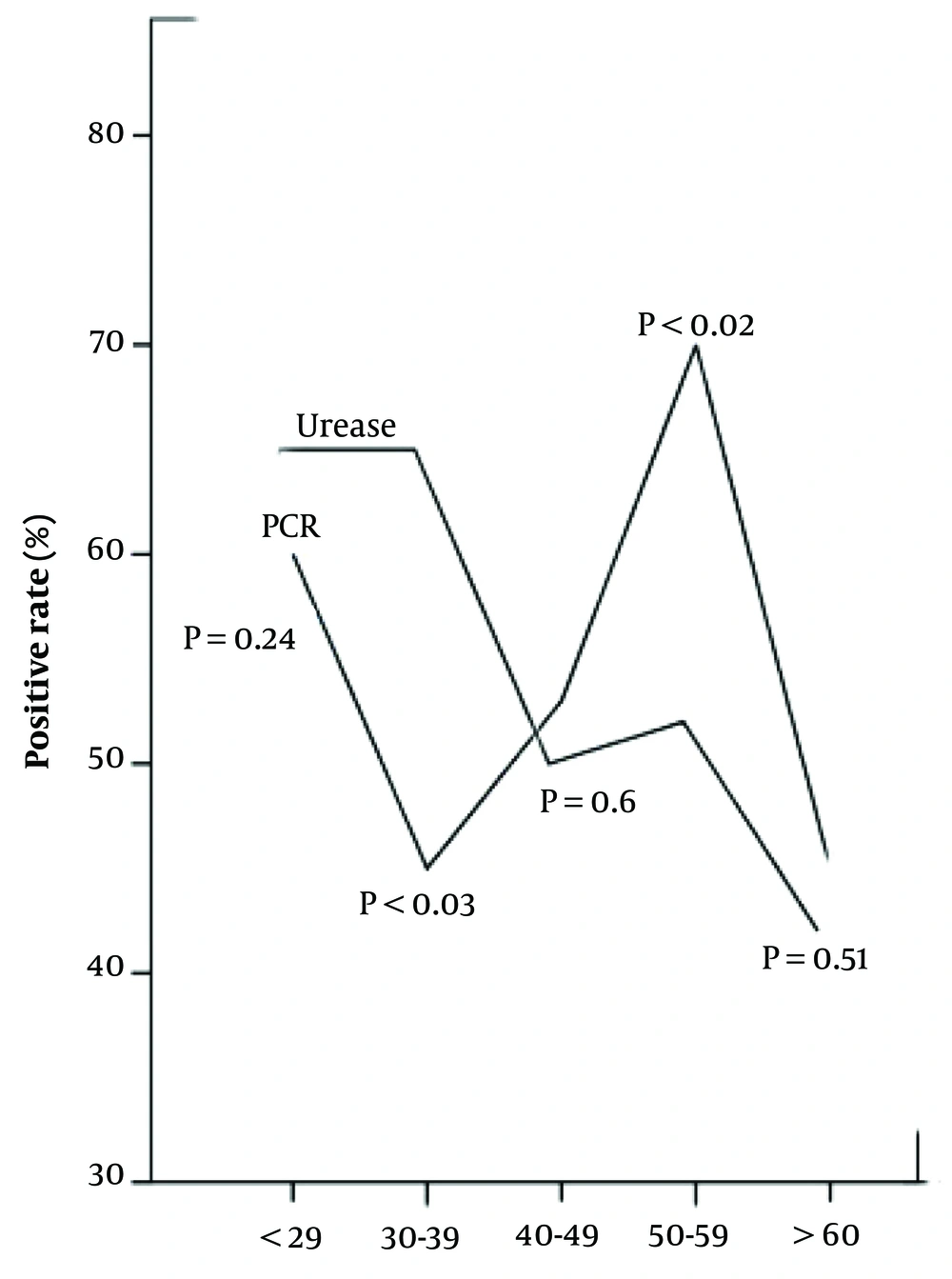
The RUT experiments showed 58% positive samples in the total of 100 subjects, among which 52.5% and 47.5% were women and men, respectively. Polymerase chain reaction targeting ureC gene was used as a specific marker which plays a key role during the bacterial invasion into gastric tissues and disease pathogenesis. The PCR products were visualized by electrophoresis in 2.5% agarose gel appearing as an 119 bp amplicon (Figure 1). With high consistency with the RUT results, the PCR findings demonstrated 57% positive cases for H. pylori infection, suggesting RUT as a sensitive method to screen for the infection in suspected people. H. pylori positive cases measured by RUT and PCR with age, sex and job of the subjects were studied. There is no significant interaction between PCR and RUT-positive subjects and social-geographic (Figure 2). Furthermore, when the patients were divided into two groups according to PCR/urease positive, higher prevalence of the group PCR positive was seen in 50 - 59 ages (Figure 3). However, urease positive showed a significantly higher prevalence in 30 - 39 ages (Figure 3). In fact the prevalence of urease positive is further in younger patient, and its prevalence would be less in older ages. Hence, there is a significant difference between urease and PCR positive at the middle and old age. Table 1 shows the prevalence of H. pylori infection according to gender. Among 100 patients, 64 (64%) of female were found to be H. pylori-positive (0.04). There was difference in prevalence of H. pylori infection according to gender. However, between H. pylori positive samples according to RUT and PCR has not seen significant difference in female and male subjects. The frequency distribution of the most common symptoms showed that the majority of subjects suffered from abdominal pain (29%), gastric burning (8%), and constipation (14%). In some other cases, they complained from a combination of several symptoms (Table 2). To confirm identity of the amplified PCR products as ureC gene, the amplified products were sent for sequencing. Following purification procedure, the sequence of the products were determined and aligned in silico using NCBI BLAST tool. The results revealed a high similarity between the obtained sequences with previously annotated sequence in the NCBI database. Owing to the fact that the biopsy specimens were collected from human gastric tissues, the alignment results must indicate no sequence of human DNA. Of note, no human DNA template was shown to be aligned with the query sequence obtained from our experiments, suggesting a highly specific PCR results. The amplicon was shown to have the following sequence:
5ˊGGGGATATGGGAACATTGGGGCCTTTAAGGATA CCTAAATCCCAAGATTTAGAATTGAAGCATTGCGCGATT GGGGATAAGTTTGTGAACAATTTTCTTTGAGGTTTTC3ˊ
| Test | ||||
|---|---|---|---|---|
| Urease | UreC Gene | |||
| Positive | Negative | Positive | Negative | |
| Gender | ||||
| F | 33 | 20 | 35 | 21 |
| M | 24 | 23 | 23 | 21 |
| Symptoms | H. pylori Positive, Frequency (%) | ||
|---|---|---|---|
| PCR | Urease | PCR/Urease | |
| Abdominal pain | 31 | 29 | 27 |
| Gastric burning | 9 | 8 | 10 |
| Abdominal pain and constipation | 15 | 14 | 15 |
| Abdominal pain and weakness and fatigue | 5 | 5 | 4 |
| Abdominal pain and severe weight loss | 5 | 5 | 5 |
| Abdominal pain and cramps | 6 | 6 | 8 |
| Gastric pain | 6 | 6 | 6 |
| Abdominal pain and gastric burning | 8 | 7 | 7 |
| Abdominal pain and nausea | 3 | 3 | 2 |
| Others | 18 | 17 | 20 |
Abbreviation: PCR, polymerase chain reaction.
5. Discussion
Numerous studies have supported the damaging effects of Helicobacter pylori infection in human gastrointestinal system causing various disorders such as gastritis, gastric and duodenal ulcers (1, 16). The bacterium was also shown to be involved in development of gastric cancer (3). Various epidemiologic investigations have reported variable worldwide prevalence of this infection from 40% in developed countries to even 90% in some developing countries (8). Having various universal reports and even the inter provincial differences, urged us to have a provincial look at H. pylori infection in Qazvin, Iran. Polymerase chain reaction is a very useful advanced tool used to detection of microorganisms in various tissues. However, the results of studies that used PCR to detect oral H. pylori were again highly variable, with detection rates ranging from 0 to 90%. Lu et al. (17) evaluated five different primers in the PCR method for the detection of H. pylori DNA which ureC gene PCR was the most sensitive and specific for detecting H. pylori in gastric biopsy samples. Consistently, we used ureC gene in PCR methods and reported a high prevalence of the infection in this province using PCR and RUT tests as two specific methods detecting the presence of H. pylori. In the present study, PCR gave the accuracy results highest and precision compared with the other method used. This explains that PCR is able to detect H. pylori DNA even if the bacterium is present in very few numbers. According to our results and the results of others, we can conclude that the PCR technique is highly sensitive and specific, and is regarded the method of choice for detecting H. pylori DNA in the oral cavity. Moreover, PCR technique requires minimal biopsy material as compared with fair-sized specimens that are needed for culture and histology to ensure successful detection. In the current study, a total of 100 individuals suffering from symptoms of upper gastrointestinal disorders were studied in regard to possible H. pylori connection. Our results indicated 58% of the patients were positive for H. pylori, which mostly agree with previous global results. The prevalence of H. pylori infection was reported in Kargar et al. study to be 94.31% in Chaharmahal Bakhtiari province, Iran (18). Rostami-Nejad et al. studied H. pylori prevalence in Tehran, Iran in 2007 by analyzing biopsy specimens of 450 patients using serologic assays (19). They showed 91% positive samples in the participants. Similar study conducted by Shokrzadeh et al. using histologic analysis revealed 86.8% out of 303 subjects to be positive for the infection in Tehran (20). During the years of 2000 - 2002, Mitipat et al. studied biopsy specimens of 110 individuals with gastrointestinal symptoms using culture techniques and RUT experiment and showed 58% positive H. pylori infections in Thailand (21), which highly agrees with our findings in Qazvin. Furthermore, another study by Nwodo et al. in Kaduna, Kaduna State, Nigeria studying 225 sera samples of patients with gastritis and peptic ulcers using ELISA; showed 80.4% prevalence of H. pylori (22). Another survey analyzing the H. pylori prevalence in Pakistan indicated 26.9% positive samples out of 4700 patients during 5 years of study (23). In Australia also, Windsor et al. reported the overall prevalence of H. pylori infections to be 76% using urease breath test (UBT) in 520 suspected individuals (24).
Several interfering factors including socio-economic conditions such as low incomes, populated families, and low public health levels were reported to affect H. pylori prevalence in developing countries (10). The prevalence of infection has been also shown to vary within the different ethnical groups in a certain country (9). Additionally, some studies have globally reported a tight association between H. pylori infection and age. According to these researches, it was demonstrated that H. pylori infections grow by age; which is mostly due to the unfavorable life style and a poor healthcare situation (25). However, our study showed no significant correlation in this regard. Consistent with our study, many previous studies also ruled out the correlation between age and H. pylori infection (20, 21, 26-28). It was speculated that family relationships may play roles in transmission of the infection from adults to children. It is also argued that the higher prevalence in adults may not be due to the direct effect of age but it can be due to the higher chance of exposure to the bacteria during a longer life span. It was also proposed by Jahan et al. that the dissimilar prevalence among different populations would be due to the cultural, genetic and life style differences (25).
Despite a bit higher infections in women, our study indicated no statistically significant association between sex and the chance of developing the infection. However, as it was previously shown by Hestvik et al. (29) and Shi et al. (26) this relationship was significant. Some former studies such as those conducted by Shi et al. found a significant correlation between education and career, with H. pylori infection (26); while we have rejected any relationship between these factors in our study. In agreement with our data, this correlation was also denied by Shokrzadeh et al. (20) and Mitipat et al (21).
5.1. Conclusions
Taken together, our study reported a high prevalence of H. pylori infections among men and women in Qazvin province, Iran in the patients with upper gastrointestinal disorders. However, the prevalence was shown to be independent of the participant’s age, sex, and their career. A comprehensive regional statistics of H. pylori infection in Iran can pave the way toward a more favorable and standardized healthcare system.