1. Background
More than 200 foodborne illnesses are transmitted to humans, among which one of the most severe diseases is listeriosis, caused by consuming food contaminated with Listeria monocytogenes (1, 2).
Listeria is one of the four major foodborne pathogens identified by the WTO (1). Milk, being one of the most consumed and perishable animal foods, is highly vulnerable to contamination by Listeria, particularly L. monocytogenes (3, 4). Among high-risk groups such as neonates, pregnant women, immunocompromised individuals, and older people, listeriosis has a hospitalization rate of 91%, with a fatality rate of 50% among neonates and a mortality rate of 20 - 30% (5, 6).
Of the six Listeria species studied, L. monocytogenes is the most prevalent and antibiotic-resistant food pathogen. The pathogenicity of L. monocytogenes is influenced by various virulence factors, including actin (actA), internalins (in1A, in1B, in1C, and in1J), phosphatidylinositol phospholipase C (PI-PLC, plcA), listeriolysin O (hlyA), virulence regulator (prf A), and invasion-associated protein (iap) (7, 8). These bacteria can survive in both extracellular and intracellular environments, transitioning from a saprophytic to a pathogenic stage via a complex network of several regulators such as prf A, σB, and CodY (9, 10).
There are several Listeria pathogenicity islands, i.e., LIPI-1 to LIPI-4, but the genes that determine the potential virulence of isolates (especially prf A and hly genes) are present on LIPI-1 (5, 6, 11).
Listeria monocytogenes possesses numerous regulatory genes and transport proteins that enable it to colonize diverse ecosystems (7, 8).
2. Methods
A descriptive cross-sectional study was conducted to examine the prevalence of L. monocytogenesin dairy products and to evaluate the prf A gene for its virulence and pathogenicity.
2.1. Sample Collection
This study involved collecting 100 dairy product samples from Ardabil city, Iran. The samples included n = 60 milk samples, n = 35 cheese samples (n = 15 soft cheese samples and n = 20 plain cheese samples), and n = 5 samples from infected animals.
2.2. Sample Preparation, Processing and Strain Isolation
The samples were prepared for processing. Milk samples were filtered to remove impurities, and 20 mL was taken for further analysis. Cheese samples were carefully crushed, and 20 g was taken for testing. Similarly, samples from infected animals were filtered, and 20 mL was taken for processing. The samples were processed in the laboratory according to ISO 11290-1:2017 (https://www.iso.org/standard/60313.html, accessed on 1 January 2018) with slight modifications (9). The double enrichment technique was employed for the isolation of Listeria species. Twenty grams of each sample were introduced into brain heart infusion (BHI) broth (12), Listeria enrichment broth (3), and Mueller Hinton agar. The samples were then incubated at 4℃ for 7 days with shaking at 100 - 120 rpm in a shaker incubator and adjusted to 0.5 MacFarland before handling (12, 13).
The inoculum was streaked onto agar plates and incubated at 37°C. After incubation, colonies were selected for secondary enrichment at 30°C for 24 hours. Finally, these colonies were streaked onto fresh agar plates and incubated at 37°C (14).
2.3. Identification and Characterization of Listeria Species Pluralis
The isolated bacterial colonies were screened for identification and characterization using various biochemical tests, including Gram staining, catalase test, oxidase test, motility test at 25℃ and 37℃, methyl red/Voges-Proskauer (MR/VP) test, nitrate reduction test, citrate test, hydrolysis of esculin, fermentation of sugars (glucose, mannitol, xylose, maltose, α-methyl-D-mannoside, and rhamnose), β-hemolytic activity, and Christie–Atkins–Munch-Peterson (CAMP) test. All tests were conducted according to the guidelines provided by FDA BAM and ISO 11290 methods (15, 16).
2.4. DNA Extraction and PCR Amplification of prf A Gene
After the identification of L. monocytogenes through biochemical tests, further molecular characterization was conducted via DNA extraction followed by prf A gene amplification for evaluation purposes. The strain was molecularly identified by 16S rRNA gene sequencing. The PGA DNA extraction kit (Pouya Gene Azma Co, Iran) was used to extract genomic DNA from the Listeria isolates according to the manufacturer’s instructions. The extracted DNA was then preserved at -20℃ for PCR (17).
For the evaluation of the prf A gene in L. monocytogenes, conventional PCR was performed, which is an in vitro enzymatic amplification of the targeted DNA sequence based on oligonucleotide primer-directed DNA synthesis by DNA polymerase. A 217 base pair fragment of the prf A gene was amplified using the routine PCR method (18). The primers LIS-R: TGA GCA ACG TAT CCT CCA GAG T and LIS-F: TCA TCG ACG GCA ACC TCG G were designed for this purpose (19, 20).
The process began with pre-incubation of DNA samples at 95℃ for 5 minutes, followed by 40 cycles of denaturation, where double-stranded DNA (dsDNA) was converted into single-stranded DNA (ssDNA) at 95℃ for 30 seconds. The next step involved hybridization or annealing of the two primers to the complementary region of the DNA template at 54℃ for 30 seconds, followed by primer extension at 72℃ for 30 seconds. The final step was elongation or synthesis of DNA from sites dictated by the primers at 72℃ for 10 minutes. Each newly formed amplified fragment served as a template for subsequent cycles, resulting in an exponential increase in the number of target DNA copies. In just 2 - 3 hours and 30 - 40 cycles, up to 106 copies of the target DNA were amplified.
All amplified samples were run on 1% agarose gel for gel electrophoresis and visualized as bands on the gel using a DNA stain (21).
2.5. Enumeration of Listeria monocytogenes
After the molecular and biochemical characterization, the CFU/mL of L. monocytogenes was analyzed. All the positive milk samples were subjected to serial dilution to determine the colony-forming units per milliliter in each sample. A 10 μL culture was suspended in 10 mL of BHI broth, incubated at 37°C for 24 hours, and then subjected to serial dilution. Then, 1 mL of the sample from an appropriate dilution was taken and subjected to the pour plate and spread plate methods. The CFU/mL was measured by counting the colonies at specific dilutions (21, 22).
Listeria monocytogenes isolates from dairy products and infected animals were tested for antibiotic susceptibility. The disc diffusion method was used to evaluate the effectiveness of various antibiotics against these bacteria, following the guidelines of the Clinical and Laboratory Standards Institute (13). A well-isolated colony of L. monocytogenes was transferred to 10 mL of BHI broth and incubated at 37°C for 24 hours, then diluted 1:10 in 9 mL of 0.1% peptone water to achieve a 0.5 MacFarland standard. The diluted culture was spread on Mueller-Hinton agar plates, and antibiotic discs were placed on the plates (3, 14).
The antibiotics used for testing included ampicillin, kanamycin, penicillin G, co-trimoxazole, tetracycline, ciprofloxacin, cephotaxime, erythromycin, imipenem, and chloramphenicol. The agar plates were then incubated at 37°C. To determine the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC), various dilutions of antibiotics were prepared, and the isolates were cultivated to identify the concentration at which no visible growth was observed (1).
2.6. Congo Red Phenotype Detection
For detecting the pathogenicity and virulence of L.monocytogenes, it was also subjected to the Congo red agar method. All the isolates were tested for the production of exopolysaccharides by streaking them onto Congo red agar plates and incubating them at 37℃ for 24 hours under aerobic conditions, followed by incubation at room temperature for another 24 hours. The biofilm-forming ability was assessed based on media discoloration and colony morphology. The results were considered positive when the isolates displayed colonies in a black to brown, wrinkled, or matte form (23).
3. Results
The results of the current study are as follows:
The collected samples were analyzed for the presence of Listeria spp. A cross-sectional descriptive analysis was employed to determine the prevalence of Listeria spp. across different sample types. Descriptive cross-sectional statistics provide an overview of the data, offering a clear picture of the attributes of the analyzed population at a specific point in time. Here, the total number of samples analyzed and the corresponding positive results, along with percentages, are presented, as frequencies and percentages are the most common ways to describe categorical data.
Among 60 milk samples, 5 (8.3%) contained Listeria spp. About 9 (25.7%) of the cheese samples tested positive for Listeria spp., while 5 (100%) of the samples collected from infected isolates had Listeria spp., as shown in Table 1. Overall, of the 100 total samples, 19 tested positive, indicating a 19% prevalence of Listeria spp. However, 55 milk samples and 26 cheese samples (plain cheese + soft cheese) tested negative for Listeria spp.
| Types of Samples | No. of Samples Analyzed | Positive Results for Listeria Species Pluralis |
|---|---|---|
| Milk | 60 | 5 (8.3) |
| Cheese (plain cheese+soft cheese) | 35 (20 + 15) | 9 (25.7) |
| Infected animal isolates | 5 | 5 (100) |
| Total number | 100 | 19 |
Processing of the Collected Samples
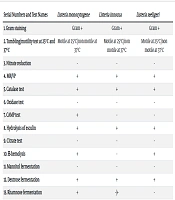
The collected isolates from all the samples were subjected to biochemical testing to identify and classify the different Listeria spp. The results confirm and verify the presence of L. monocytogenes, L. innocua, and L. seeligeri, as shown in Table 2.
| Serial Numbers and Test Names | Listeria monocytogene | Listeria innocua | Listeria seeligeri |
|---|---|---|---|
| 1. Gram staining | Gram + | Gram + | Gram + |
| 2. Tumbling/motility test at 25℃ and 37℃ | Motile at 25℃/non motile at 37℃ | Motile at 25℃/non motile at 37℃ | Motile at 25℃/non motile at 37℃ |
| 3. Nitrate reduction | - | - | - |
| 4. MR/VP | + | + | + |
| 5. Catalase test | + | + | + |
| 6. Oxidase test | - | - | - |
| 7. CAMP test | + | - | - |
| 8. Hydrolysis of esculin | + | + | + |
| 9. Citrate test | - | - | - |
| 10. Β-hemolysis | + | - | + |
| 11. Mannitol fermentation | - | - | - |
| 12. Dextrose fermentation | + | + | + |
| 13. Rhamnose fermentation | + | -/+ | - |
| 14. Xylose fermentation | - | - | + |
| 15. α-methyl-D-mamoside | + | + | -/+ |
| 16. Phosphatidylinositol phosphatase | + | - | - |
Biochemical Testing
After the biochemical characterization of L. monocytogenes, DNA was extracted and amplified by PCR. To confirm the identity and virulence of L. monocytogenes, the prf A gene was amplified. The frequency of the prf A gene observed in L. monocytogenes was approximately 94.73% (n = 18), indicating that L. monocytogenes strains possess the prf A gene responsible for their pathogenicity.
The results show that out of 19 positive samples, 14 milk samples contained L. monocytogenes, 4 cheese samples contained L. innocua, and 1 sample from an infected animal contained L. seeligeri. The corresponding percentages are presented in Table 3.
| Name of Isolates | Sample | No. (%) |
|---|---|---|
| Listeria monocytogene | Milk | 14 (73.6) |
| Listeria innocua | Cheese | 4 (21) |
| Listeria seeligeri | Animal isolates | 1 (5.26) |
| Total | - | 19 |
Species Identified from the Contaminated Samples
The isolated samples of L. monocytogenes were then enumerated, and the CFU details are provided in Table 4.
| No. of Samples | CFU/mL |
|---|---|
| 1. | 320 |
| 2. | 460 |
| 3. | 570 |
| 4. | 690 |
| 5. | 780 |
| 6. | 940 |
| 7. | 1520 |
| 8. | 1690 |
| 9. | 1840 |
| 10. | 1920 |
| 11. | 2140 |
| 12. | 2250 |
| 13. | 2380 |
| 14. | 2530 |
Listeria monocytogenes (CFU/mL) of in All 14 Positive Milk Samples
The enumeration of L. monocytogenes in milk samples indicates a wide range of microbial loads. As the sample numbers progress, the CFU/mL values also increase. The variation in results reflects the heterogeneity in the level of contamination of L. monocytogenes in the dairy samples. After determining the CFU/mL of L. monocytogenes in the dairy samples, the antimicrobial resistance profile of L. monocytogenes was assessed, as shown in Table 5.
| Variables | Dose (μg/disc) | Sensitivity of the Isolates (%) | ||
|---|---|---|---|---|
| R | I | S | ||
| Ampicillin | 10 μg | 1 (7.14) | 5 (35.71) | 8 (57.14) |
| Canamycin | 10 μg | 5 (35.71) | 1 (7.14) | 8 (57.14) |
| Penicillin G | 10 U | 4 (28.57) | 5 (35.71) | 5 (35.71) |
| Co-trimoxazole | 25 μg | 6 (42.85) | 3 (21.42) | 5 (57.14) |
| Tetracycline | 30 μg | 7 (50) | 3 (21.42) | 4 (28.57) |
| Ciprofloxacin | 5 μg | 2 (35.71) | 1 (7.14) | 11 (78.57) |
| Cephotaxim | 30 μg | 8 (57.14) | 4 (28.57) | 2 (18.18) |
| Erythromycin | 15 μg | 3 (27.27) | 3 (18.18) | 8 (57.14) |
| Imipeneme | 10 μg | 4 (27.27) | 2 (14.28) | 8 (57.14) |
| Chloramphenicol | 30 μg | 5 (27.27) | 3 (27.27) | 6 (42.85) |
Antimicrobial Resistance Profiles of Listeria monocytogenes Isolates
The antimicrobial resistance, MIC, and MBC of L. monocytogenes were determined from both food-based and animal source samples. The list of antibiotics and their effects on L. monocytogenes are shown in Table 5. Ciprofloxacin exhibited the highest efficacy, while cephotaxime showed both sensitivity and resistance. The MIC and MBC of Ciprofloxacin were determined to be 4.15 and 8.5, respectively.
The use of Congo red agar further confirmed the presence of L. monocytogenes with the prf A virulent gene, as the change in color and biofilm formation indicated its presence. The results showed that 100% of isolates from infected animal sources tested positive with the Congo red test, while among food samples, 78.57% (n = 11) yielded positive results.
4. Discussion
The results indicate that milk has 8.3%, cheese has 25.7%, and infected animal isolates have 100% contamination from Listeria species (1). These findings show that 7.67% of Listeria species are found in milk and related products (10). Similarly, one study stated that about 8.5% of Listeria species were found in milk samples and 6.8% in cheese (8).
PCR provides sensitive and rapid detection of L. monocytogenes in food products (24). The current study indicates a prevalence of the prf A gene (94.73%) in L. monocytogenes via PCR. A similar study using PCR detected virulence genes (prf A, hlyA, actA, and inlA) in L. monocytogenes isolated from contaminated milk samples (24). Another study also supports the prevalence of the prf A gene in L. monocytogenes isolates extracted from various sources such as the environment, milk, and infected animal samples analyzed via PCR (14). Another study in Italy supports these findings, showing that the prf A gene and act A gene were 100% dominant in L. monocytogenes (15). A previous study conducted in Nigeria aligns with the current study, stating that only three genes (hly A, iap, and prf A) are virulence-associated genes in L. monocytogenes (15).
The current study found that among analyzed dairy product samples, milk had 73.6% (n = 14) L.monocytogenes, cheese had 21% (n = 4) L. innocua, and infected animal isolates had 5.26% (n = 1) L. seeligeri. These findings are supported by a previous study (1), which reported that 2.28% of L. monocytogenes was found in milk samples, 9.09% in bulk milk tanks, 2.85% in cheese, and 1.82% in infected cow milk (1). Another study exhibited the presence of L. monocytogenes in cheese and related processing facilities, though with a low prevalence, consistent with the current findings (10).
The presence of L. seeligeri in milk, feed, feces, water, and the environment and the presence of L. innocua in milk, feces, feed, the environment, and water have been documented previously (25). Another study supporting our findings reported that L. monocytogenes was found in milk (13%), udder swabs (19%), and cattle fecal samples (43%). Prior studies also mention that the prevalence rate of L. monocytogenes in raw cow milk can vary from 0% to more than 45% (21).
Another corresponding study shows that raw milk contains 25% Listeria species, among which approximately 2% of the isolates are L. monocytogenes and 5% are L. innocua (22). Another prior study reports about 5.90% occurrence of L. monocytogenes in raw milk (18, 26).
The prevalence of L. monocytogenesvaries among different food sources due to several influencing parameters (2), including geographical region, season, type of sample, time, processing environment, farm size, transport conditions, isolation techniques, management practices, milking process, storage methods, silage quality, sample size, and the level of sanitation in food processing and manufacturing facilities (2, 27, 28).
Approximately 60% of foodborne outbreaks are attributed to bacterial biofilm formation (26). One of the most common biofilm-forming, foodborne pathogens is L. monocytogenes, which is responsible for the spoilage of dairy and ready-to-eat products (20). The current study indicates biofilm formation verified by Congo red agar in L. monocytogenes isolated from food samples (78.5%) and from infected animal samples (100%). Previous studies also confirm the importance of the prf A gene in biofilm formation in L. monocytogenes (29, 30).
4.1. Conclusions
This study found that dairy products (milk, cheese) and samples from infected animals were contaminated with Listeria species. Molecular analysis confirmed the presence of L. monocytogenes and the virulence gene prf A. The study highlights the importance of prf A in the pathogenicity and biofilm formation of L. monocytogenes.