1. Background
A brain tumor is an atypical lesion in the brain tissue that is caused by the abnormal growth of brain cells and is one of the most common neurological diseases that affects more than 300,000 people worldwide every year (1). Because of limited cranial space (2), brain tumors are fundamentally dangerous due to its invasive nature (3). Brain tumors can be fatal especially malignant types (4).
Brain tumors or intracranial tumors (5) can be included in malignant (cancerous) or benign (non-cancerous). Brain tumors are divided into primary and secondary types. Primary tumors originating from brain cells and nerve cells are gliomas, meningiomas, pituitary tumors, etc. Secondary tumors, or metastases, are tumors that reach the brain from cancer in other parts of the body and form a mass (6). Also, tumors can have different symptoms, such as headaches, vision problems, and hearing disorders, based on their location in the brain tissue (7).
There are different methods and steps to diagnose brain tumors. In the first stage, doctors examine the patient's nerves and the balance and coordination between the organs and then they use imaging methods such as CT and MRI to examine the tumor and its location. Due to the high dose of X-rays in CT imaging and its relatively low contrast, the MRI technique is preferable (8). Then, doctors (such as radiologist, neurologist, neurosurgeon and etc.) can determine whether the tumor is benign or malignant by sampling. Chemotherapy and surgery are the most common treatment options that can be used to treat brain tumors. Surgery may be ultimately successful in cases where the brain tumor is benign. One of the essential criteria to accurately assess the effectiveness of new treatments for brain tumors is the growth dynamics determined by MRI. However, manual measurement of tumor diffusion in high-contrast MRI scans is prone to error and leads to relatively different results. Traditional diagnoses can have disadvantages and errors, and due to the invasiveness of sampling to detect the type of tumor, artificial intelligence techniques have been advanced (9).
Artificial intelligence (10) techniques are formed on the idea that systems can learn from suitable data, recognizing patterns, and decisions making with minimal hominid involvement. Machine learning (ML) (10) is defined as one of the subfields of artificial intelligence that is skilled of autonomous learning to automatically modify and better the performance index of a task based on its previous tasks. Machine learning relies on various algorithms to resolve data problems, in which the nature of algorithm used depends on a series of features, including the category of problem, the amount of variables, and the most suitable model (11). In line with the studies of Fabelo et al. (12), the support vector machine (SVM) algorithm has been used in this project. According to the available data, this algorithm can be used both linearly and non-linearly. Support vector machine classifiers work well in high-accuracy spaces and use much less memory. Support vector machine is used to create the best decision line or boundary to separate the N-dimensional space into classes to easily place the new data point into the correct category in the future (13).
Considering the importance and pervasiveness of brain tumor disease, a correct and timely diagnosis of the disease can reduce mortality worldwide and prevent its progression. Various studies have been conducted on brain tumor diagnosis and classification with different algorithms (7, 11-18).
2. Objectives
The innovation of the present research is the combined use of morphological features and discrete wavelet transform (DWT) to design an automatic system. The presented algorithm can be used as a diagnostic tool to increase the accuracy of robotic surgery and brain tumor classification.
3. Methods
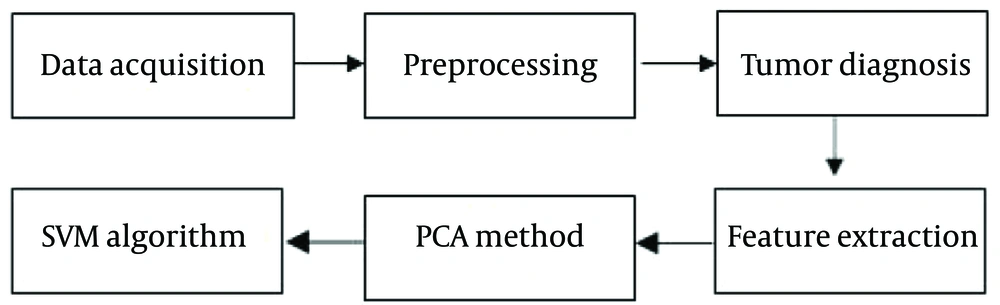
The steps of the current research are shown in Figure 1 below:
3.1. Data Acquisition
In this retrospective study, 160 T2-weighted MR images were used for brain tumor diagnosis and classification. The images were obtained from https://kaggle.com and consisted of 140 malignant tumors and 20 benign tumors. The data was divided into two groups: Benign tumors and malignant tumors. The following steps were performed on the images:
Preprocessing: Medical images, including MRI, often contain noise. Filters, such as the median, ideal, Butterworth, and wavelet filters, were used to process the images and remove noise.
- The median filter is a non-linear filter that eliminates salt and pepper noise from MRI images (19).
- The Fourier ideal filter removes specific frequencies from the MRI image while leaving other frequencies intact (20).
- The Fourier-Butterworth filter removes low-frequency components from the MRI image, preserves the high-frequency components, and makes the edges of the MRI image stand out (21).
- A wavelet filter analyses MRI images at different scales and resolutions (22).
Various criteria, such as mean square error, signal-to-noise ratio, and pixel signal-to-noise ratio, are used to compare and evaluate filter performance (23).
3.2. Tumor Diagnosis
In this study, a tumor is defined as a tissue whose strength or coherence is at least 50% of the brain tissue. The strength of the tumor tissue is determined by its density and area. “Solidity” as a morphological operations is used to segment or separate the tumor tissue. Solidity is the ratio of pixels in the objects to pixels of the convex hull image. For clearness, the convex hull of the objects is characterized in dark gray into the images (24). In fact Solidity gives a measure of the compactness of the object. Objects with high solidity (78.99) more likely to be tumor (25). These operations are mathematical tools used to extract image features. In this study, morphological operations are used to describe the general shape of a complication or region or to reveal the boundaries of the image (26).
3.3. Feature Extraction
856 morphological and wavelet features were extracted from benign and malignant brain tumor images and stored in two separate files by using radiomic (15).
Radiomic features can be separated into five groups: Size and shape based-features, descriptors of the image intensity histogram, descriptors of the relationships between image voxels (e.g. gray-level co-occurrence matrix (GLCM), run length matrix (RLM), size zone matrix (SZM), and neighborhood gray tone difference and etc. A complete explanation of texture features for radiomics can be found in Parekh and Jacobs (27). The feature vectors are extracted as a matrix with n * m sizes. The columns of the matrix of feature vectors (n) correspond to the number of images (160), and the rows of the matrix (m) are the variables of the feature vector (856) .The labeling process has been shown in Appendix 1. The feature matrix is extracted from the images by applying the DWT to the tumor area (28). The feature matrix's performance in classifying tumor images will be compared.
3.4. Principal Component Analysis Method
The principal component analysis (PCA) method (29) is used to reduce the dimensions of a matrix of feature vectors. In this method, the rows of the feature vector matrix represent images, while the columns represent variables. Principal component analysis aims to reduce the number of variables or columns in the matrix while keeping the same number of rows. This method transforms the original correlated variables into new, uncorrelated ones called principal components. The covariance matrix method is one of the ways to calculate the essential components.
To summarize the PCA method, the average of each column in the feature vector matrix is subtracted from that column's elements to get a zero-based data matrix. The covariance matrix is then obtained, followed by the eigenvalues and eigenvectors of the matrix. The sorted particular values are then isolated, and the first N essential components are kept while the rest are removed. This results in a more minor dimension of image data by multiplying the remaining components in the original data (X).
3.5. The Support Vector Machine
Support vector machine algorithm (30) is used to classify brain tumors into benign and malignant categories. The training data D (MRI images) is assumed to be a set of n points. In this study, training data (n = 128) is given as Equation 1:
Equation 1.
Equation 2.
In this relation,
Equation 3.
Equation 4.
3.6. Calculation of the Accuracy of the Image
To evaluate the accuracy of an image classification algorithm, the cross-validation method is used. This involves dividing the total number of positive and negative images in the database into k groups. In this case, 160 benign and malignant tumor images are divided into k-5 groups. Sixteen benign tumor data and 112 malignant tumor data are used for network training, while four and 28 malignant tumor data are used for network validation. In each process step, one group is used as a set of test images, while the other K-1 groups are used as training images. The classification algorithm trains the SVM using the training images and then calculates the accuracy parameters of the classification algorithm using the test images. By averaging the accuracy parameters of these five steps, the overall accuracy parameters of the classification algorithm are obtained. Appendix 3 shows how the algorithm works (31, 32). This method provides an effective way to check the accuracy of the image classification algorithm.
In a binary classifier (with two possible classes), based on the classification output, there will be the following four states, which are displayed in the confusion matrix (33):
- True positive = images that were positive (a malignant tumor) and correctly predicted positive.
- True negative = images that were negative (benign tumor) and correctly predicted negative.
- False positive = images that were positive and predicted as negative.
- False negative = images that were negative and falsely predicted as positive.
The higher the number of true positives and true negatives, and the lower the number of false positives and false negatives, the better the algorithm's performance.
These criteria are obtained from the Equation 5 (33).
Equation 5.
4. Results
The results of using median, ideal, Butterworth and wavelet filters on a sample image are included in the Appendices 4-7 with explanations.
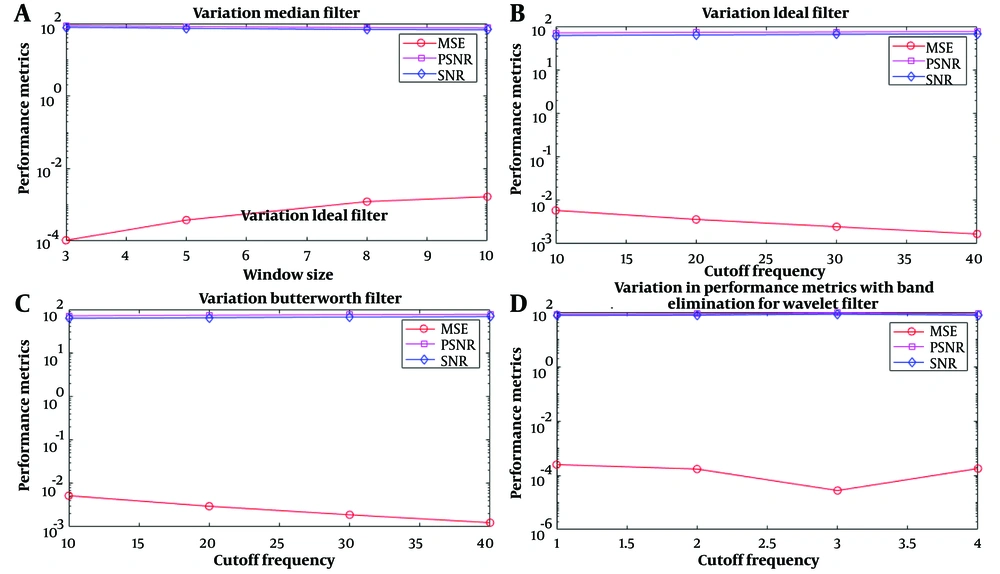
The results of applying filters for MRI data preprocessing are shown in Figure 2. Filters have been compared regarding signal-to-noise ratio (SNR), RSNR, and MSE performance at different frequencies. Finally, the best filter will be selected to apply brain tumor MRI data preprocessing. The higher the SNR, the better the characteristic because more helpful information will be received in the form of a signal than unwanted information or noise. MSE measures the amount of error in statistical models. It is always a positive value that decreases as the error approaches zero. PSNR is the peak signal-to-noise ratio, a term between a signal's maximum possible power and destructive noise's power. In the absence of noise, MSE is zero, and PSNR is infinite.
Shows the performance evaluation results of filters with MSE, PSNR, and signal-to-noise ratio (SNR) criteria. According to the result, PSNR and SNR were unsuitable parameters to show filter performance because they changed according to one feature. According to A, as the median filter window size increases, the filter's performance becomes weaker; in B and C, with the increase in the cut-off frequency of the Ideal-Butterworth filter, the MSE measure increases linearly with a low slope and has the same performance; in D, by increasing the cut-off frequency, the wavelet filter performs better up to the frequency range of 3 and then changes linearly.
According to the figures and results obtained from applying the filters, the median filter with a window size of 3 and the wavelet filter with a cut-off frequency of 3 had the best performance on the imported MRI images.
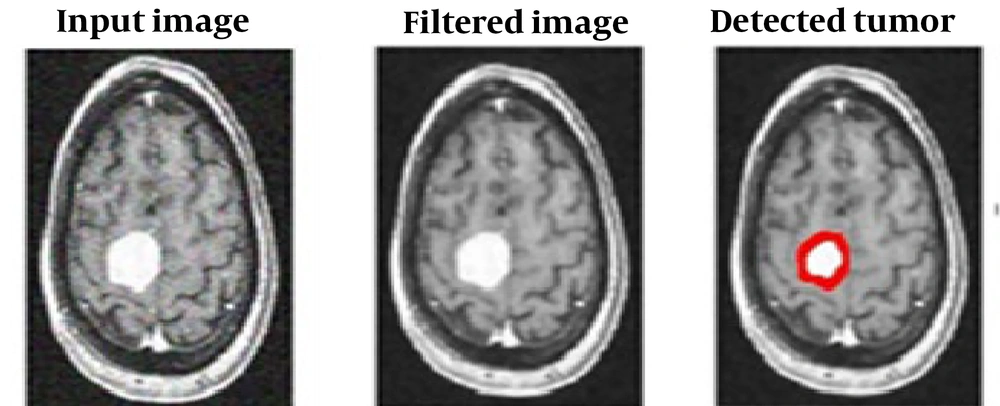
After passing the images through the appropriate filter, with the correct thresholding in the area and strength of the tumor tissue, the tumor is diagnosed and isolated. Morphology operations have been applied to this semi-processed image, and information about the strength and areas of acceptable tumor locations has been obtained. The result of separating the tumor from the main brain tissue is shown in Figure 3.
In this study, 160 features of MRI data have been extracted from MRI images using DWT features, and the number of features extracted is equal to 856, which are mean, average, contrast, strength, complexity, etc. Using the PCA method, these features have reached 19. Finally, the SVM algorithm has succeeded in classifying brain tumor into two types: Benign and malignant. The results of the confusion matrix are shown in Table 1.
| SVM | Benign | Malignant |
|---|---|---|
| Benign | 17 | 3 |
| Malignant | 5 | 135 |
According to the table above, using the DWT feature transformation and PCA method and the number of correct detections (135 + 17), the accuracy, precision, recall, and specificity of the SVM algorithm in this study were 95%, 88%, 91%, 91%. According to these results, the proposed method can be used in different fields of medicine.
5. Discussion
Different types of tumors can be benign or malignant. This study examined two types of brain tumors, benign and malignant, from 160 MRI samples. The necessity of the current study was based on the fact that brain tumors are one of the most common neurological diseases that can endanger the lives of people of all ages. A timely and accurate diagnosis of the tumor and its type and the prescription of fast and correct treatment can save many people's lives. Also, in medical robotic surgeries, it can help correct tumor diagnosis and increase the accuracy of surgery. This paper has automated the analysis method for brain tumor detection through image processing techniques. This research focuses on four directions. On the one hand, preprocessing data and images includes operations such as removing noise using different filters and comparing them. The findings of this study showed that the median filter with a window size of three and the wavelet filter with a cut-off frequency of three had the best performance among the selected filters in removing noise from brain tumor MRI images. Kavya et al. (34) disclosed that using the median filter increases the filtering and removes the noise or artifact of medical images, which was consistent with our results. In the second direction, the segmentation method has been used for brain tumor MRI images after improving image quality and reducing noise.
In the aspect of diagnosing and separating tumors from brain tissue, in addition to using morphological operations and relying on age, gender, brain structure, disease center, and similar cases, it is very focused on accurate classification, area, and strength, according to studies (17) and with regarding the size of the tumor, in this study, the tumor is a tissue whose density is at least 50% different from the brain tissue, which is different according to the type of tumor and the size of the tumor (35), which can be related to the inherent characteristics of images. In the third direction, the features of MRI images are extracted using the DWT method, according to the study of Arora et al. In a research study, 1024 features were extracted from MRI images using DWT. In accord with our methods, later, these features were reduced to 7 using the PCA method (36). Additionally, the PCA method was utilized in this study to decrease the quantity of features.
One of the contributions of this paper is to propose a method that combines various techniques to identify normal and abnormal MR brains. By using the SVM algorithm, the brain tumor has been successfully divided into two types: Benign and malignant. Another study by Kalam et al. (16) used the ANFIS and SVM algorithms to detect and isolate the tumor. The results of that study is similar to us. Similarly, in the study by Anantharajan et al. (14), the SVM algorithm is used to classify brain tumors which is in line to our results.
This research can enhance the quality of medical images and accurately diagnose the location and type of brain tumor by employing a high-precision artificial intelligence system in robotic medicine and surgery.
5.1. Conclusions
Our results present a completely automatic segmentation using SVM for segmentation and classification brain tumors. The tumor regions designated are spatially small and consistent regarding image content and provide a suitable and strong guide for the resulting segmentation and classification. Our proposed method can attain promising tumor segmentation in combination with supervised approachs. Our experimental results indicate that the proposed method will help identify the exact position of the brain tumor correctly and fast.
The investigational outcomes showed 95% accuracy of detecting tumor and normal tissues in magnetic resonance images. The findings conclude that the suggested approach may be sufficient for primary diagnostic for radiologists or any clinical and technical experts specially robotic surgeries technician or engineer.