1. Background
Pseudomonas aeruginosa is a pathogenic bacterium that is gram-negative (1). Pseudomonas aeruginosa is responsible for various infections, including pneumonia, surgical site infections, urinary tract infections, and bacteremia. It is estimated to have a prevalence of 7.1% - 7.3% among all patients (2). Keratitis is an ophthalmological condition characterized by inflammation of the cornea, leading to potential impaired vision in severe instances. This disease can arise from microbial infection, including bacteria, fungi, viruses, or protozoans, as well as non-infectious factors like eye trauma, exposure to chemicals, or ultraviolet light (3). One of the bacteria that causes keratitis is P. aeruginosa. Pseudomonas aeruginosa demonstrates adaptability to unfavorable conditions in hosts by secreting several virulence factors, which play a crucial role in achieving effective infection and inducing disease (4). Lipopolysaccharide (LPS) serves as a crucial surface structural element that safeguards the external leaflet and poisons host cells. The lipid A in LPS exhibits endotoxicity, which facilitates tissue injury, attachment, and detection by host receptors. The LPS has the potential to be associated with antibiotic resistance and the production of biofilms (5, 6). Furthermore, the development of biofilms is linked to drug resistance, which is attributed to the flagellum, pili, and other adhesins. In addition, there exist six distinct secretion systems, including flagella (related to the T6SS), pili (associated with the T4SS), and the multi-toxin components type 3 secretion system (T3SS). These systems play crucial roles in several biological processes such as host colonization, adhesion, swimming, and swarming, as they respond to chemotactic signaling (7). The presence of T3SS exoenzymes was identified as significant virulence factors that trigger P. aeruginosa keratitis. The T3SS is a specialized protein export system that establishes a needle-shaped complex between bacterial and host cells to facilitate the transportation and release of four exotoxins, namely exotoxin S (exoS), exoT, exoU, and exoY (8). The exoS is a cytotoxin consisting of 453 amino acids (48 kDa) that demonstrates the ability to activate both GTPase-activating protein (GAP) and ADP ribosyl transferase (ADPRT) enzymes (8).
Protease IV (piv), sometimes referred to as lysyl endopeptidase, is a 26 kDa iron-regulated enzyme that belongs to the chymotrypsin family of proteins. It is encoded by the prpL gene. The activation of the host immunological defense system is initiated by piv via the degradation of fibrinogen, plasminogen, and immunoglobulin G, as well as the inactivation of complement components, including C3 and C1 (9). The corneal pathogenicity of P. aeruginosa has been associated with several virulence factors, one of which is the synthesis of the extracellular enzyme piv. The correlation between tissue damage during keratitis and the production of this enzyme has been demonstrated in many animal models (10, 11).
Addressing the escalating issue of antibiotic resistance is an important challenge that afflicts contemporary civilization (12). The multifaceted nature of the antibacterial action employed by P. aeruginosa poses challenges in effectively eliminating the bacteria from healthcare environments. Pseudomonas aeruginosa’s outer membrane has several porins that regulate cellular mobility, as well as efflux pumps that actively expel antimicrobial agents. In addition, compared to other gram-negative bacteria, it exhibits lower permeability to antibiotics (13).
It is known that resistance and virulence genes are also transmitted simultaneously. To achieve substantial advancements in addressing the global issue of antibiotic resistance and virulence, it is imperative to adopt a novel approach that acknowledges the interconnectedness of their regulation and transmission. The facilitation of gene expression, transfer, and regulation is enhanced not only when both genetic elements are co-located on a plasmid but also when bacteria exist in a biofilm state (14, 15). By determining the relationship between different virulence genes and antibiotic resistance, it is possible to find strategies to prevent the further transmission of these genes together.
2. Objectives
Therefore, this study aimed to investigate exoS and piv genes among P. aeruginosa resistant strains isolated from patients with corneal keratitis.
3. Methods
3.1. Sampling and Culture
The ethics committee and institutional review board of Islamic Azad University Sanandaj Branch granted approval for this study, which adhered to the principles outlined in the Declaration of Helsinki. An analysis of all patients with a diagnosis of corneal ulcer at Imam Khomeini Hospital (Kermanshah, Iran) was performed. All corneal ulcers, with the exception of peripheral infiltrates less than 3 mm (as they are less likely to be caused by microbial keratitis or require cultures) and without any indication of corneal thinning upon slit lamp examination, underwent corneal scraping. Corneal scrapes were acquired subsequent to the administration of topical anesthesia. Corneal samples were obtained using a sterile scalpel blade under the observation of a slit lamp biomicroscope. Capillary ulcer samples were utilized for the direct inoculation of culture media, specifically MacConkey and Cetrimide agar. The plates were then incubated at a temperature of 37°C for 48 hours. The P. aeruginosa strains were identified via microbiological tests (Catalase test) (16) to detect the presence of the enzyme catalase, which breaks down hydrogen peroxide. Positive results (bubbles) indicate the presence of catalase.
3.2. Antibiotics Susceptibility Testing
DNA was extracted using the DNA extraction kit (Sinaclon, Iran) according to the manufacturer's instructions. Antibiotic susceptibility was confirmed by the disk diffusion technique on Muller-Hinton medium (Merck, Germany), performed according to the Clinical Laboratory Standard Institute (CLSI) guidelines. Paper disks were impregnated with antibiotics (Padtanteb, Iran): Imipenem (10 μg), Piperacillin (100 μg), Ciprofloxacin (5 μg), Tobramycin (10 μg), Ceftazidime (30 μg), Gentamicin (10 μg), Ticarcillin (75 μg), Amikacin (30 μg), Cefixime (5 μg). They were incubated overnight at 37°C. The diameter of the zone of inhibition was measured, and the results were interpreted as sensitive, intermediate resistant, or resistant, based on CLSI guidelines (17).
3.3. Virulence Gene Detection
The PCR technique was employed to confirm the existence of the exoS and piv genes, utilizing the primers and conditions outlined in Table 1. Each P. aeruginosa isolate was subjected to screening for the presence of virulence genes in a 20 μL reaction volume PCR assay, employing the Master mix (Sinaclon, Iran), and adding 0.4 μM of forward and reverse primers per gene to the amplified, nuclease-free water and extracted genomic DNA. The amplification process was conducted using a Labcycler (Sensoquest, Germany) with the following conditions: 94°C for 3 minutes, followed by 35 cycles of amplification at 94°C for 30 seconds, 60°C for 60 seconds, 72°C for 60 seconds, and a final step of extension at 72°C for 7 minutes. Agarose gel electrophoresis was performed on the PCR products using a 1.5% agarose gel in a 1x concentrated TBE buffer (Sinaclon, Iran). The electrophoresis was conducted at 80 V for a duration of 1 hour using a 50 bp DNA ladder (Sinaclon, Iran). Subsequently, the DNA bands were observed and measured using UV light.
| PCR Product | Sequence (5′ → 3′) | Amplicon Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| exoS | 118 | 62 | |
| Forward | GCGAGGTCAGCAGAGTATCG | ||
| Reverse | TTCGGCGTCACTGTGGATGC | ||
| piv | 600 | 60 | |
| Forward | GCTTCAGTTTGGTCAGGAGC | ||
| Reverse | CAGGCTCATTGAGTGTGGAA |
Abbreviations: exoS, exotoxin S; piv, protease IV.
3.4. Statistical Analysis
In this research, the data were entered into SPSS software (SPSS Inc., Chicago, Illinois, USA) version 22. Statistical analysis was performed using logistic regression. In addition to the P value, logistic regression also provides us with odds ratio data. This data shows the strength of the correlation between the independent and dependent variables.
4. Results
4.1. Samples and Resistance Pattern
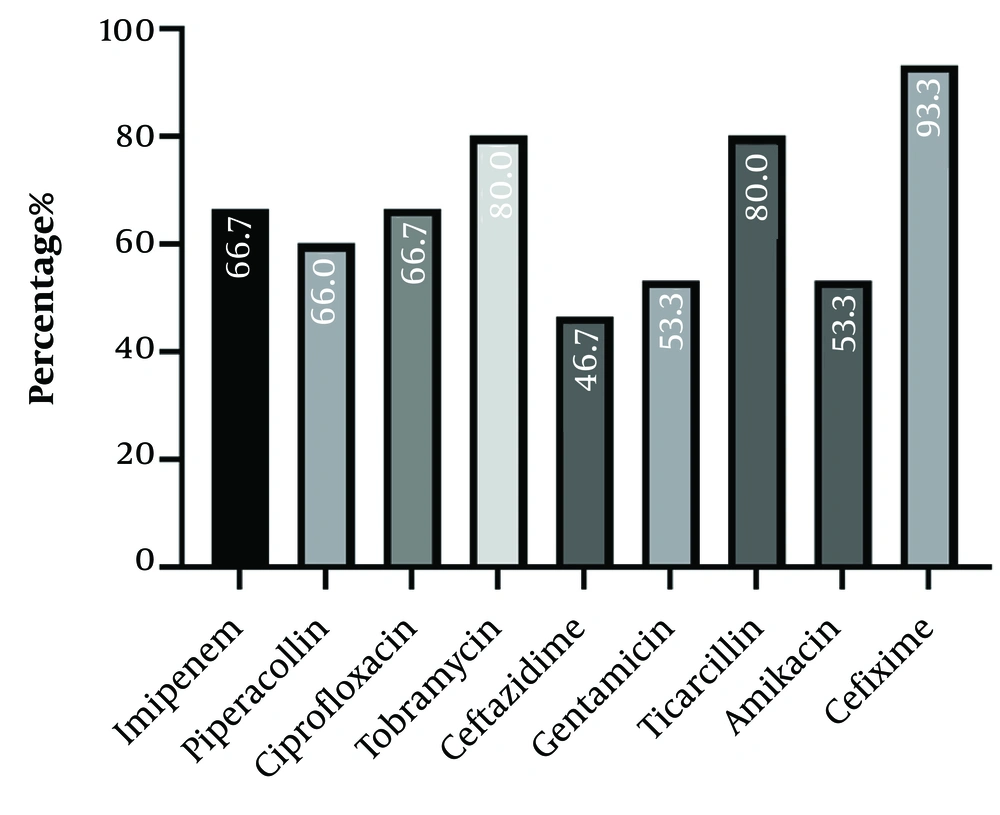
A total of 175 corneal cultures were conducted. The patients had an average age of 58.4 ± 19.3 years, and 114 (65.14%) were male. Out of the entire set of ocular specimens, 72 isolates (41.14%) exhibited positive culture results. Among culture-positive isolates, 15 (20.83%) were identified as P. aeruginosa via biochemical tests. Figure 1 displays the distribution of antibiotic resistance profiles. The antibiotic resistance rates of P. aeruginosa were found to be highest against cefixime (93.3%) and lowest against ceftazidime (46.7%).
4.2. Relationship Between Exotoxin S and Protease IV Genes and Antibiotic Resistance
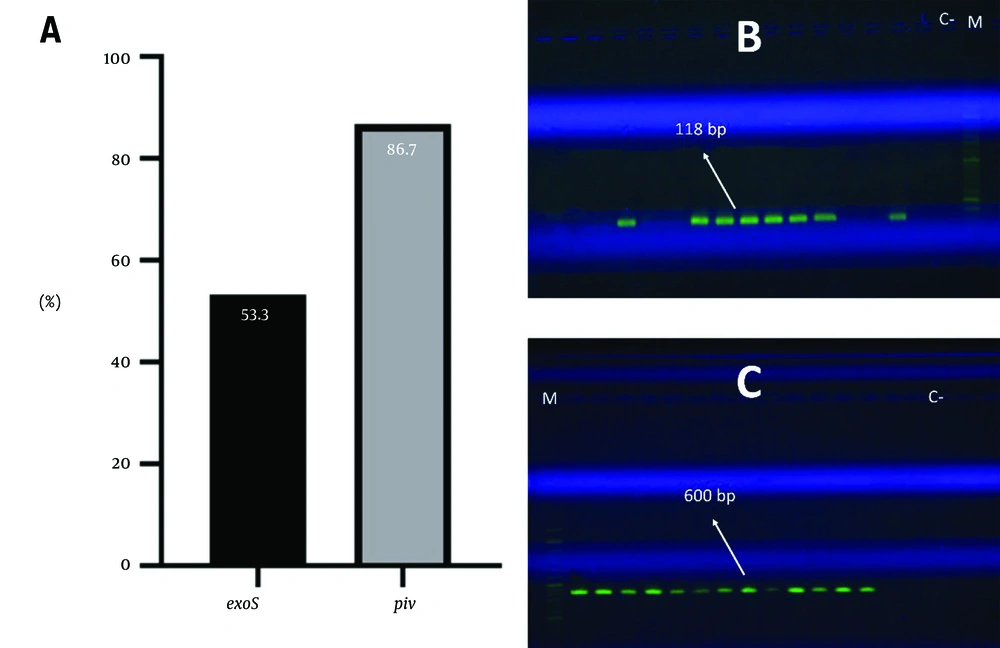
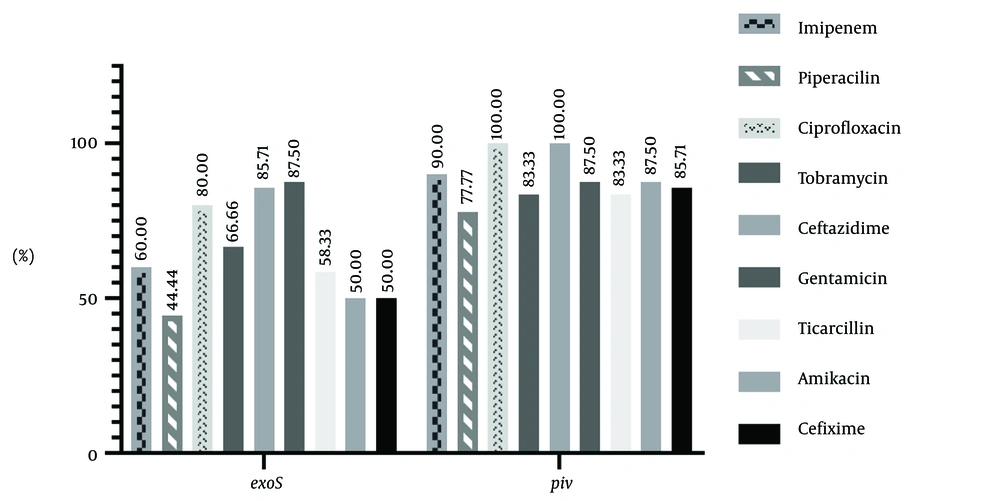
The results showed that 53.3% of the strains have the exoS gene and 86.7% have the piv gene (Figure 2). This study also examined the correlation between the existence of virulence genes (exoS and piv) and antibiotic resistance. The frequency of genes associated with antibiotic resistance in strains is depicted in Figure 3. Table 2 presents the statistical analysis conducted using logistic regression. The analysis of the table reveals that there is no significant correlation between the existence of the piv gene and antibiotic resistance (P > 0.05). Regarding the exoS gene, it is noteworthy that a significant correlation exists between the gene's existence and resistance to ceftazidime (P = 0.033, OR: 18, CI = 1.267 - 255.744) and gentamicin (P = 0.014, OR: 42, CI = 2.136 - 825.715). Strains that are resistant to ceftazidime and gentamicin are more likely to possess the exoS gene, indicating a direct association between the exoS gene and resistance to these antibiotics.
| Antibiotic Resistance | P-Value | OR | CI (95%) | |
|---|---|---|---|---|
| Lower | Upper | |||
| Imipenem | ||||
| exoS | 0.468 | 2.250 | 0.251 | 20.131 |
| piv | 0.598 | 2.250 | 0.111 | 45.723 |
| Piperacillin | ||||
| exoS | 0.403 | 0.4 | 0.47 | 3.424 |
| piv | 0.999 | - | - | - |
| Ciprofloxacin | ||||
| exoS | 0.999 | - | - | - |
| piv | 0.999 | - | - | - |
| Tobramycin | ||||
| exoS | 0.999 | - | - | - |
| piv | 0.999 | - | - | - |
| Ceftazidime | ||||
| exoS | 0.033 | 18 | 1.267 | 255.744 |
| piv | 0.999 | - | - | - |
| Gentamicin | ||||
| exoS | 0.014 | 42 | 2.136 | 825.715 |
| piv | 0.919 | 1.167 | 0.059 | 22.937 |
| Ticarcillin | ||||
| exoS | 0.448 | 2.8 | 0.196 | 40.057 |
| piv | 0.999 | - | - | - |
| Amikacin | ||||
| exoS | 0.782 | 0.75 | 0.98 | 5.768 |
| piv | 0.919 | 1.167 | 0.059 | 22.937 |
| Cefixime | ||||
| exoS | 0.999 | - | - | - |
| piv | 0.999 | - | - | - |
Abbreviations: OR, odds ratio; CI, confidence interval; exoS, exotoxin S; piv, protease IV.
5. Discussion
Bacterial keratitis is a prevalent condition affecting the corneal tissue, posing a significant risk to vision. The occurrence of endophthalmitis and subsequent eye loss can be attributed to the ineffective or delayed treatment of this prevalent infectious eye disease. Bacterial keratitis is caused by a significant multitude of microorganisms. Previously, Streptococcus pneumoniae was the most frequently found pathogen in bacterial corneal ulcers. However, the prevalence of Pseudomonas and Staphylococcus infections has risen due to the growing use of contact lenses (20, 21). Pseudomonas aeruginosa is a globally significant pathogen that exhibits a notable degree of genetic variation. Most isolates have virulence genes that are involved in the production of pyocyanin, phospholipase C, piv, alkaline protease, elastase B, and some exoenzymes (22). The virulence feature of the P. aeruginosa strain is a multifaceted subject. The genotype makeup of a particular strain may be a significant factor in determining its varying pathogenicity potential and complex drug-resistance spectrum, particularly in a specific subgroup of strains. The results showed resistance to imipenem, piperacillin, ciprofloxacin, tobramycin, ceftazidime, gentamicin, ticarcillin, amikacin, and cefixime in 66.7%, 60%, 66.7%, 80%, 46.7%, 53.3%, 80%, 53.3%, and 93.3%, respectively. The 93.3% cefixime resistance in P. aeruginosa is highly unusual, suggesting a combination of intrinsic and acquired resistance mechanisms. While P. aeruginosa is known for its inherent resistance, the specific high resistance to cefixime likely involves a combination of its restricted outer membrane permeability, efflux pumps, and potentially acquired beta-lactamases (23).
In the study conducted by Alamri et al., the resistance rates were as follows: 33.3% for aztreonam, 36.7% for imipenem, 16.7% for meropenem and ceftazidime, 13.3% for ciprofloxacin, and 10% for gentamicin and cefepime (24). Based on the differences in the type of clinical samples and geographical region, variations in the resistance pattern of the strains can be observed. Mirzaei et al. conducted a study in Iran which documented resistance rates of 64%, 52%, 58.7%, and 62.7% to imipenem, gentamicin, ceftazidime, and ciprofloxacin, respectively (25). The findings of the Mirzaei et al. study exhibit a higher degree of similarity to the current study due to the shared geographical context of Iran. If there is a difference in the results, it can be due to the difference in the type of sample (25). A multiple-drug resistance (MDR) rate of 86.66% is a cause for concern, even when aligned with regional data. Potential biases in the data may include differences between hospital-acquired (nosocomial) and community-acquired strains, as well as sampling and testing methodologies.
Corneal epithelial cells exhibit varying effects of T3SS effectors based on their genotypes. Strains possessing the exoS genotype have the ability to infiltrate and proliferate within corneal epithelial cells, both in vitro and in vivo. The survival of P. aeruginosa within corneal epithelial cells was revealed to be dependent on the presence of a functioning T3SS (8). The pathogenicity of P. aeruginosa in experimental keratitis has been attributed to the type III effector ExoU, which has been found to trigger fast lysis of epithelial cells and macrophages. Cytotoxic strains, namely exoU+ strains, have the ability to harm the epithelia on a corneal surface that is not damaged, as long as there is prolonged contact with the bacteria. ExoU+ strains may exhibit more adhesion under contact lens wear circumstances (26).
The correlation between tissue damage during keratitis and the production of piv has been demonstrated in many animal models. Research has demonstrated that piv exhibits the ability to degrade a diverse range of host proteins, encompassing host defense components such as complement and IgG class immunoglobulin (10). The presence of ring abscess lesions in pseudomonal keratitis has been found to be associated with piv (27). Therefore, the isolated strains had a high prevalence of piv (86.7% of the piv gene).
While piv has been shown to be important for pathogenesis, studies indicate that there's no significant correlation between its presence and antibiotic resistance. This means that the loss or presence of piv does not directly impact an organism's ability to develop resistance to antibiotics (28). The prevalence of the exoS gene in the present study was 53.3%. The frequency of antibiotic resistance and virulence genes can vary in each geographical region. In other studies conducted in Iran, the incidence of exoS was shown to be higher. In a study conducted by Fazeli and Momtaz (29), the incidence of exoS in isolates from Iranian hospital infections was reported as 67.64%. Dadmanesh et al. (30) published a rate of 73.91% for exoS. Additionally, Shokrzadeh et al. (31) found that the frequency of the exoS gene in Sari, Iran was 77.8%. The elevated incidence of exoS observed in our investigation may be attributed to the substantial clonal variability exhibited by the isolates. The lower frequency of exoS in the present study could be because the studied strains had a more exoU+-exoS- profile.
Numerous investigations have been conducted to examine the potential association between the virulence and resistance spectrum of P. aeruginosa, yielding inconsistent findings. Previous reports have shown a positive correlation between the exoU+ genotype of P. aeruginosa and a moderate resistance phenotype, as well as a negative correlation with extended-drug resistance (XDR). Additionally, it has been demonstrated that the exoU+ genotype is more likely to be resistant to fluoroquinolones when there is a gyrA mutation and overexpression of the efflux pump (32).
In P. aeruginosa, there's a relationship between the virulence factor exoS and the development of carbapenem resistance. While not directly causing resistance, exoS, along with other virulence factors like exoY and exoT, can contribute to the pathogenicity of P. aeruginosa, particularly in carbapenem-resistant strains. Moreover, exoS expression is often co-expressed with other virulence factors like exoU, and this co-expression can be linked to enhanced virulence in carbapenem-resistant strains (33). In contrast to the present study, some previous studies reported that the exoS genotype has a relation to the phenotype of carbapenem resistance, suggesting that exoS may be incongruous with genes associated with carbapenemase production to a certain degree in clinical strains (32, 34, 35). They reported that given the intricate regulatory network regulating virulence and drug resistance in P. aeruginosa, it is possible that the feasible antagonism between exoS and carbapenem resistance could confer advantages to the bacteria, enabling them to achieve prolonged survival and infection.
In the present study, there was no significant relationship between the exoS gene and the phenotype of resistance to imipenem (carbapenem). This difference can be related to the fact that only imipenem was used as a carbapenem antibiotic and also the type of samples is different. In some bacteria, a potential antagonism, or conflict, exists between the presence of the virulence gene exoS and carbapenemase genes. Specifically, exoS was found to be less frequently detected in carbapenem-resistant Pseudomonas aeruginosa (CRPA) strains carrying carbapenemase genes, suggesting an incompatibility between the two. This finding hints at a complex relationship between virulence and resistance in these bacteria (32).
In our study, a significant relationship was observed between the presence of the exoS gene and resistance to ceftazidime (cephalosporin) and gentamicin (aminoglycoside). The utilization of antivirulence strategies, which involve combating bacterial infections by targeting virulence factors — the components that enable bacteria to cause disease — rather than directly killing the bacteria themselves, has emerged as a novel approach to solving the issue of resistant bacterial strains (22). Through the analysis of the distribution and function of virulence factors in bacteria, it is feasible to identify chemicals that can effectively hinder their activity. Moreover, by establishing a correlation between virulence genes and antibiotic resistance, the simultaneous use of antivirulence agents and antibiotics that exhibit resistance can be avoided.
Biofilm formation, though unmeasured in our study, can significantly impact gene-resistance links. Biofilms, dense microbial communities, promote horizontal gene transfer (HGT), facilitating the rapid spread of resistance genes within and between microbial populations. This can lead to increased antibiotic resistance and a broader range of resistance mechanisms compared to planktonic (single-cell) growth (36).
The main limitations of our work were the small number of available samples, which limits statistical power and generalizability, and the lack of mechanistic insights into how exoS might drive resistance to ceftazidime/gentamicin.
The strains of P. aeruginosa isolated from keratitis patients exhibit high antibiotic resistance, with 86.66% of them showing MDR. This finding underscores the need to identify novel drug compounds for these bacteria and modify antibiotic consumption patterns (e.g., stewardship strategies) within this specific geographic region. Keratitis strains mostly possess the piv gene, highlighting the importance of this virulence gene in their pathogenicity. The production of exoS in strains also has a prevalence of 53.3%. The findings of our study demonstrate a significant correlation between the exoS gene and individual resistance to ceftazidime and gentamicin agents. This link should be taken into account when analyzing the outcomes of infection. No significant correlation between the piv gene and antibiotic resistance was observed. Further research with larger sample sizes and in-depth investigation of the mechanism is warranted in the future. Finally, the identification of piv and exotoxin inhibitors can also provide assistance. For future investigations, biofilm assays are suggested for a deeper exploration of the correlation with resistance.