1. Background
Salmonella spp. is a gram-negative, coccobacillus, facultative anaerobic, non-acidic, and non-spore-forming organism with flagella and the diameter of 2 - 4.5 micrometers. These microorganisms belong to the Enterobacteriaceae family and have biochemical and serological diversities. Salmonella spp. has more than 2,500 different serovars and is divided into three distinct species, including Salmonella typhi, Salmonella choleraesuis, and Salmonella enterica (1, 2).
Salmonella enterica serovar typhimurium is among the three most important serovars of salmonella in the world. Salmonella infection occurs in humans in the form of food poisoning, gastroenteritis, typhoid fever, and sepsis in some cases (3, 4). The pathogenesis of salmonellosis encompasses their survival and proliferation within macrophages. Sip and sop proteins, which are produced by the type III secretory system (T3SS), are the most important factors involved in the pathogenesis of this organism (5).
Several reports have been published regarding the epidemics of enteritidis, typhimurium and infantis serovars in Iran (6, 7). Gastroenteritis is a highly prevalent infection caused by S. typhimurium (8). In a study conducted in Iran, the prevalence of salmonella among children with gastroenteritis was reported to be 40% (9). Today, beta-lactamases are the most significant antibiotics used in the treatment of salmonella infections. Unfortunately, the addition of antibiotics to the feed of livestock, the improper and excessive administration of these agents, and lack of proper monitoring on their use have led to the emergence of antibiotic-resistant bacterial strains (10, 11).
Antibiotic-resistant genes in Salmonella spp. induce resistance against common antibiotics. Salmonella spp. could obtain resistance to antibiotics through various mechanisms, such as chromosomal mutations, resistance transmission through genetic materials (mostly by plasmids), reduction of cell wall permeability to antibiotics, enzymatic inactivation of antibiotics, efflux pump mechanism for taking out antibiotics, and altered target position of antibiotics (4, 12). The identification of beta-lactamases in clinical specimens based on their spectrum of effect, level of enzyme production, and diversity of enzyme activity in bacteria is of utmost importance in the prevention and treatment of the associated infections in the community.
Several studies have investigated the resistance of Salmonella spp. to extended-spectrum beta-lactamases (ESBLs). In Iran, reports have been published regarding the resistance of Salmonella spp. to beta-lactamases, the cause of which could be attributed to the excessive and incorrect use of antibiotics (especially in animal feed) by individuals and specialists (13, 14). The most important mechanism of resistance to these drugs is the transfer of resistance genes through plasmids, which often occurs in Enterobacteriaceae, and the expression of their effects through multiple enzymes. The identification of appropriate drug substrates for these enzymes could prevent the emergence of resistant strains and their prevalence, as well as treatment failure and substantial treatment costs (10, 15-18).
2. Objectives
The present study aimed to determine the antibiotic resistance pattern of S. typhimurium isolated from the clinical samples of children and identify antibiotic resistance genes (blaTEM, blaSHV, blaCTX-M, blaPSE-1, sipB/C, and cmlA/tetR) in the isolates using multiplex PCR.
3. Methods
3.1. Sample Collection and Isolate Identification
This cross-sectional, descriptive study was conducted on 110 fecal samples collected from children with diarrhea who were referred to the hospitals in Tabriz, Iran during eight months. The patients were selected via random sampling. Data were collected on their age, gender, and hospitalization/outpatient status. The inclusion criterion of the study was the manifestation of symptoms such as abdominal pain, diarrhea, vomiting, and fever. Bacterial samples were cultured on Salmonella Shigella agar (SS) and xylose lysine deoxycholate (XLD) agar.
At the next stage, suspected colonies of Salmonella spp. were inoculated to selenite F (SF) broth, and the isolates were identified using common biochemical and microbiological tests, such as urea, Simon citrate, triple sugar iron (TSI), citrate, and methyl red/Voges-Proskauer. In addition, serotyping was performed to determine somatic (O), flagella (H), and capsular antigens (Vi) using monovalent antisera and the slide agglutination method. The mentioned media were purchased from Merck Company, Germany, and S. typhimurium ATCC 14028 was used as the positive control.
3.2. Antibiogram Test and Double-disc Synergy Test (DDST)
The antibiotic resistance pattern of the isolates was evaluated using the Kirby-Bauer method in accordance with the CLSI instructions. Briefly, a 0.5 McFarland concentration of bacteria was prepared in broth. After culturing the bacteria on the Mueller-Hinton agar (MHA), the resistance of the isolates to the antibiotics discs was assessed. The antibiotics included tetracycline (30 μg), chloramphenicol (30 μg), cotrimoxazole (25 μg), ciprofloxacin (5 μg), cefotaxime (30 μg), ceftazidime (30 μg), nalidixic acid (30 μg), ceftriaxone (30 μg), gentamicin (10 μg), cefepime (30 μg), amikacin (30 μg), ampicillin (10 μg), aztreonam (30 μg), and imipenem (10 μg) (MAST, UK) (19). In addition, the ESBL-producing isolates were identified using the combined disc method, with the discs containing ceftazidime (30 μg), cefotaxime (30 μg), ceftazidime/clavulanic acid (30 μg/10 μg), and cefotaxime/clavulanic acid (30 μg/10 μg) (Himedia, India). After incubation at the temperature of 37°C for 24 hours, the growth inhibition zone diameter of ≤ 5 millimeters around the ceftazidime/clavulanic acid discs were compared with the ceftazidime disc. Moreover, the growth inhibition zone diameter of ≤ 3 millimeters around the cefotaxime/clavulanic acid discs was compared with cefotaxime (20).
3.3. Identification of blaTEM, blaSHV, blaCTX-M, blaPSE-1, sipB/C, and cmlA/tetR Genes in the Isolates
In the present study, the bacterial genome was extracted using a commercial DNA extraction kit (Invitek Strateg Business, Canada), and the target genes were identified using specific primers and multiplex PCR (Table 1).
| Target Genes | Sequence (5’→3’) | Product Size (bp) | References |
|---|---|---|---|
| blaTEM | 861 | (21) | |
| F | GAGTATTCAACATTTCCGTGTC | ||
| R | TAATCAGTGAGGCACCTATCTC | ||
| blaSHV | 747 | (22) | |
| F | ATGCGTTATATTCGCCTGTG | ||
| R | TGCTTTGTTATTCGGGCCAA | ||
| blaCTX-M | 592 | (23) | |
| F | ATGTGCAGYACCAGTAARGTKATGGC | ||
| R | TGGGTRAARTARGTSACCAGAAYCAGGG | ||
| sipB/C | 232 | (24) | |
| F | ACAGCAAAATGCGGATGCTT | ||
| F | GCGCGCTCAGTGTAGGACTC | ||
| cmlA/tetR | 260 | (24) | |
| F | CGCTCCTTCGATCCCGT | ||
| F | GCTGCGTTCATCTACAACAGAT | ||
| blaPSE-1 | 132 | (24) | |
| F | TTTGGTTCCGCGCTATCTG | ||
| F | TACTCCGAGCACCAAATCCG |
Primers Used to Identify Target Antibiotic-resistant Genes in Salmonella typhimurium Isolates of Children with Diarrhea in Tabriz, Iran
A PCR reaction was performed in the volume of 25 microliters, including 6.5 microliters of a PCR master mix 5X (Sinaclone, Iran; containing 3 μL of Mgcl2, 0.4 μL of dNTP Mix, 0.7 μL of primer, 0.05 U/μL of Taq polymerase enzyme, and 0.9 μL of DNA per sample) and 12 microliters of sterile deionized water. The thermal cycling program was as follows: one cycle of initial denaturation at 94°C for four minutes, 30 cycles of denaturation at 94°C for 30 seconds, annealing at 57°C for 35 seconds, and extension at 72°C for 90 seconds.
The final extension was carried out at the temperature of 72°C for 10 minutes, and the PCR products were observed after electrophoresis on 1% agarose gel compared to the standard strains of S. typhimurium ATCC 14028 and S. enteritidis ATCC 19271.
Data analysis was performed using Chi-square in SPSS version 20 at the significant level of P < 0.05.
4. Results
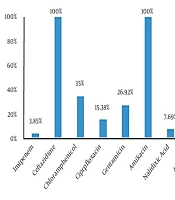
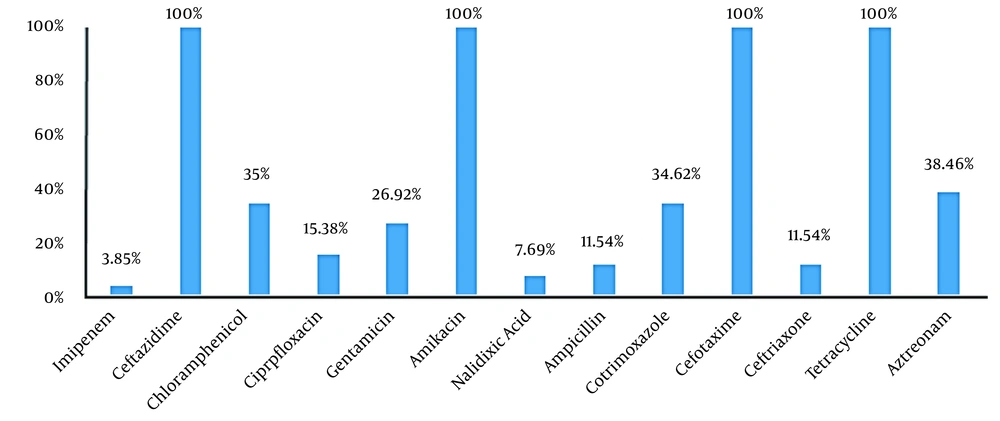
Out of 110 fecal samples, 26 cases (23.63%) were positive for S. typhimurium. In total, 16 isolates (61.54%) and 10 isolates (38.46%) belonged to the male and female patients. in addition, 76.92% of the isolates (n = 20) were collected from the inpatient ward, and 23.08% (n = 6) were obtained from the outpatient ward. The antibiogram results indicated that S. typhimurium had the highest resistance to ceftazidime, cefotaxime, amikacin, and tetracycline (100%), while the lowest resistance was observed against imipenem (3.85%) and nalidixic acid (7.69%). Moreover, 15.38% of the isolates were resistant to more than two classes of antibiotics (Figure 1).
Out of 26 S. typhimurium isolates, 15 cases (57.69%) were positive for ESBL production in the phenotypic test. On the other hand, 11 isolates (73.33%) and four isolates (26.67%) were detected in the male and female patients, respectively. The results of PCR analysis showed that cmlA/tetR (38.46%), blaTEM (34.61%), and blaCTX-M (26.92%) were the most common resistance gene in the isolates. Four isolates (15.38%) carried sipB, three isolates (11.53%) contained blaSHV, and two isolates (7.69%) carried blaPSE-1. In addition, blaTEM, blaCTX-M, and blaSHV were detected concurrently in two isolates (7.69%). All the S. typhimurium isolates that were phenotypically ESBL-positive carried at least one of the studied ESBL genes as well.
5. Discussion
Recently, Salmonella spp. has become widespread worldwide due to the emergence of numerous new serotypes compared to the past. Each year, salmonellosis accounts for a significant percentage of human infections, especially in children and the elderly (25). The results of the present study indicated that 26 S. typhimurium isolates (23.63%) were detected in 110 fecal samples. Furthermore, the results of the Kirby-Bauer test for different antibiotics showed that the highest resistance of the isolates was against ceftazidime, tetracycline, cefotaxime, and amikacin (100%). Meanwhile, the lowest resistance of the isolates was observed against imipenem (3.85%), nalidixic acid (7.69%), ampicillin, ceftriaxone, and cefepime (11.54%). Therefore, it could be concluded that these antibiotics may be used as the first choice in the treatment of salmonellosis.
In a study regarding quinolone-resistant salmonella, Abbasi and Ghaznavi-Rad isolated the samples from patients with diarrhea, reporting that out of 230 samples of infectious diarrhea, 21 cases (9.1%) were salmonella-positive. In the mentioned study, the highest antibiotic resistance was reported against nalidixic acid (71.4%) and tetracycline (8.42%) (7). In another research, Najafi et al. (26) isolated 14 (29.2%) strains of S. typhimurium from 163 samples, and the highest resistance of the isolates was against tetracycline (56.2%), streptomycin, and chloramphenicol (31.2%). In the mentioned study, all the isolates were sensitive to imipenem, amikacin, gentamicin, and trimethoprim/sulfamethoxazole (100%). Furthermore, the PCR results indicated that 62.5% and 16.6% of the isolates contained cmlA/tetR and sipB genes.
Pezzella et al. (27) investigated streptomycin and tetracycline resistance genes in the transposons and plasmids of Salmonella spp. isolates. According to their findings, 84% and 68% of the isolates were resistant to streptomycin and tetracycline, respectively. In addition, all the isolates were positive for tetA gene. In Korea, Yang et al. (24) investigated antimicrobial resistance in Salmonella enterica serovar enteritidis and S. typhimurium isolated from animal samples. The blaTEM and blaSPE genes were detected only in five of the 40 S. enteritidis isolates and 22 of the S. typhimurium isolates.
Zinatizadeh et al. (28) examined 60 clinical isolates of S. typhimurium, observing that all the isolates were sensitive to imipenem, gentamicin, and amikacin. In the mentioned study, the highest resistance of the isolates was observed against ampicillin (16.6%), amoxicillin (14.9%), and tetracycline (11.6%). On the other hand, the results obtained by Arjmand-Asl and Amini (29) demonstrated that all S. typhimurium isolates were sensitive to imipenem, gentamicin, and trimethoprim. Furthermore, the frequency of the slyA, stn, sopB, and phoP/Q genes in the isolates was estimated at 3.23%, 30.0%, 33.3%, and 3.43%, respectively. Yang et al. (30) also assessed the antibiotic resistance pattern of S. typhimurium isolates, reporting that 100%, 95.5%, and 86.4% of the isolates were resistant to streptomycin, sulfonamides, and tetracycline, respectively. In another research, Jahantighi and Amini (31) reported that out of 300 fecal samples, 11 samples (1.61%) were contaminated with S. typhi. Moreover, the evaluation of the resistance pattern of the isolates in the mentioned study indicated that the highest and lowest resistance was against imipenem, ceftriaxone (100%), and ofloxacin (5.54%), respectively. They also reported that seven (8.38%) and eight isolates (4.44%) carried the blaCTX-M and blaTEM genes, respectively, while two isolates (1.11%) were positive for blaSHV.
Rojas et al. (32) examined the antibiotic resistance of Salmonella spp. isolated from pigs, observing that the resistance rates of the isolates to ampicillin, chloramphenicol, and sulfamethoxazole was 17.24%, 25.28%, and 62.7%, respectively. In addition, 63.63% of the chloramphenicol-resistant isolates carried the cmlA/tetR gene, and 55.38% of the non-chloramphenicol-resistant strains contained this gene as well. In the mentioned study, only 13.33% of the ampicillin-resistant isolates were positive for blaPSE-1, and 46.66% harbored blaTEM. In another research, Amini et al. (33) reported the resistance of Salmonella spp. isolates to ampicillin (6.82%), chloramphenicol (4.80%), tetracycline (5.69%), cephalothin (4.80%), amoxicillin/clavulanic acid (5.56%), and trimethoprim/sulfamethoxazole (4.43%). Meanwhile, blaTEM was detected in one isolate (1.2%). All the isolates in the mentioned research were negative for blaPSE, and the human Salmonella spp. was resistant to ampicillin, cephalothin, chloramphenicol, tetracycline, amoxicillin/clavulanic acid, and trimethoprim/sulfamethoxazole.
In the studies conducted by Tajbakhsh et al. (34) and Li et al. (35), the prevalence of blaPSE and blaTEM in isolates was reported to be 63% and 18%, 3.36%, and 2.42%, respectively. On the other hand, Casin et al. (36) investigated multidrug-resistant S. typhimurium in humans and animals, reporting that 187 S. typhimurium isolates were resistant to ampicillin, including 86 human isolates and 101 animal isolates. Resistance to chloramphenicol, tetracycline, streptomycin, and spectinomycin, and sulfonamides was also reported in 69.5% of the human isolates and 64.8% of the animal samples. In the mentioned study, four beta-lactamase genes of blaTEM (24%), blaPSE-1 (78%), blaSHV, and blaoxa-2 (3% each) were also identified in the isolates. The discrepancies between the results of the present study and the aforementioned findings could be due to the differences in the geographical location and serotype of Salmonella spp. other factors such as age, genetics, and environment also influence the pathogenicity of bacteria. Virulence plasmids or virulence factors are not directly involved in the host-bacterial interaction, while most of these genes encode the proteins that are involved in the host-bacterial interaction. These effective proteins play a key role in the survival and proliferation of Salmonella spp. (29).
5.1. Limitations of the Study
The main limitation of our study was that we did not assess the minimum inhibitory concentration of the antibiotics and the association of different risk factors with S. typhimurium infection.
5.2. Conclusions
According to the results, the presence of ESBLs and antibiotic resistance in S. typhimurium strains and their simultaneous transfer to other strains in Tabriz was high, which highlights the importance of gaining more knowledge about the correlation between antibiotic resistance and the presence of virulence genes. Rational antibiotic therapy could effectively prevent the spread of these strains in hospitals and determine the clinical consequences of infection with ESBL-producing organisms.