1. Background
The recent emergence of multidrug-resistant Candida auris has become an international health problem during the past decade (1). C. auris is a recently discovered Candida/Clavispora clade member, which was first isolated from a female patients’ ear discharge in 2009 in Japan (2). Subsequently, C. auris has quickly gained a reputation as a remarkable nosocomial pathogen causing many infections throughout the world (3), including three confirmed cases from Iran, which were genetically distinct from the other known C. auris clades (4-6). C. auris is a global health issue because of its rapid global expansion, multidrug-resistance features, skin colonization capacity, nosocomial epidemic production with high mortality rates (7). Most infections were reported in immunocompromised and/or ill subjects suffering severe underlying diseases (8).
Even though the risk factors for Candida infections seem to be the same regardless of species, a recent study found that prolonged hospitalization and prolonged antifungal drug exposure are the most common risk factors for C. auris infection (9, 10). The ICUs have proven a fertile ground for establishing and spreading C. auris during the ongoing COVID-19 epidemic (11-16). Since most Candida species inhabit the gastrointestinal tract, C. auris' ability to remain on the skin surface is an unusual trait that may contribute to its efficient nosocomial transmission (3). As a result, limiting the further spread of C. auris requires early and accurate detection (17).
There are no known causes for the recent emergence of this yeast. According to whole-genome sequencing, C. auris isolates have undergone the geographical classification of five distinct clades, which have emerged independently (18). The inadequacy of conventional and commercially available biochemical tests to identify C. auris infections is a severe barrier to timely diagnosis and appropriate treatment (19).
There is the possibility of misinterpreting C. auris as other Candida species or non-Candida yeast species, even when utilizing commercial biochemical techniques allowing for species-level identification (20). Therefore, several DNA-based approaches were developed to detect C. auris in environmental and clinical specimens, some of which have been validated (21). Matrix-assisted laser desorption/ionization-time of flight mass spectrometry and polymerase chain reaction (PCR) are the most practical diagnostic techniques to accurately identify C. auris (17). In most underfunded mycology laboratories, the high cost of their procurement and operation remains a barrier (22). Several C. auris strains are reported with resistance to numerous antifungal categories (23).
2. Objectives
This study aimed to determine the spectrum of C. auris colonization from clinical skin swabs in immunocompromised patients hospitalized in ICU using the salinity-rich culture and C. auris-specific PCR.
3. Methods
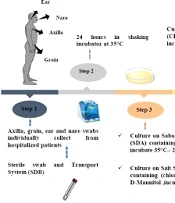
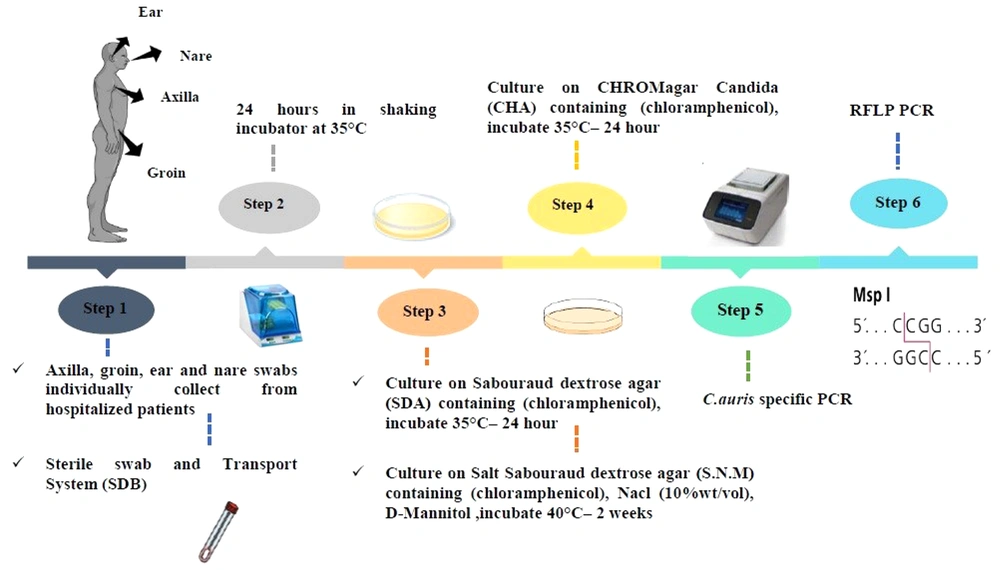
This descriptive study was conducted to evaluate C. auris colonization from the skin swabs of patients with a weak immune system or immunodeficiency in the ICUs, in Omid hospital, Isfahan, Iran, after obtaining ethical clearance by the Falavarjan Branch, Islamic Azad University, Isfahan, Iran. An informed consent was obtained from the patient or the next of kin (in the case of the unconscious patient). The active surveillance of 32 admitted patients was conducted from November 2020 to February 2021 (IR.IAU.FALA.REC.1398.046). The ICUs were the host of a mixed population of patients with immunodeficiency, including breast, intestinal, gastric, and lung tumors, leukemia, and neuroblastoma. The information, including age, gender, length of hospitalization, medical specialty, diagnosis on admission, and bathing habits of patient, were recorded upon admission (Table 1). All patients were evaluated for skin surface colonization with an expected duration of > 7 days without bathing.
| Characteristics | No. of Patients |
|---|---|
| Gender | |
| Male | 10 |
| Female | 11 |
| Child | 11 |
| Age groups | |
| < 10 | 10 |
| 11 - 19 | 2 |
| 20 - 29 | 3 |
| 30 - 39 | 2 |
| 40 - 49 | 5 |
| 50 - 59 | 7 |
| 60 - 69 | 2 |
| ≥ 70 | 1 |
| Underlying diseases | |
| Acute lymphocytic leukemia (ALL) | 8 |
| Acute myeloid leukemia (AML) | 16 |
| Lung, breast, colon, stomach and brain cancer | 7 |
| Neuroblastoma | 1 |
3.1. Clinical Samples
The skin swab samples (ie, 31 axillaries, 31 groins, two nares, one tongue, and one ear) were collected individually using the sterile swab and a transport system. Each swab was solved in 1 mL Sabouraud Dextrose Broth medium [Merck, Germany] in sterile round bottom tubes from the patients and immediately transferred to the laboratory for analysis. The swab samples were obtained by cleaning the swab in a circular motion While rotating the swab 360 degrees and applying moderate pressure to the surface. All the specimens (round bottom tubes) were put in a shaker incubator for 24 h at 35°C after receiving. Then, each tube was vortexed for 30 sec to release the specimen from the swab tip after 24 h.
3.2. Culture Conditions
About 100 μL of suspension was cultured on Sabouraud dextrose agar (SDA; Merck, Germany) supplemented with chloramphenicol and incubated at 35°C for 24 to 48 h. Then, 100 μL was cultured at the same time on Salt SDA (S.N.M) containing antibacterial chloramphenicol (0.5 g/L), NaCl (10% wt/vol), and D-Mannitol (as carbon sources) (SIGMA, Germany) and put in an incubator for two weeks at 40°C. In the next procedure, the colonies were studied, cultured on CHROMagar Candida (BioMerieux, France), and incubated at 35°C for 24 to 48 h. Finally, the plates were investigated for the color of colonies (24).
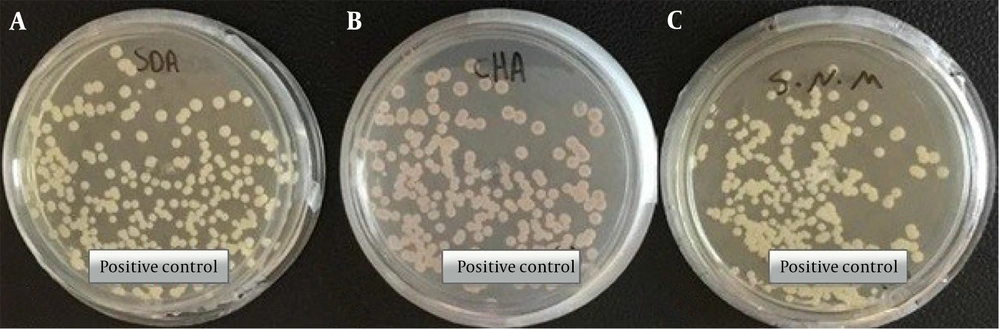
In this study, the clinical strain of C. auris (access number: MZ389242) was used as a positive control (Figure 1) (5).
Morphological characteristics of pure colonies of C. auris (MZ389242) white to cream on Sabouraud dextrose agar (A), pink on CHROMagar upon 2 days of incubation at 35°C (B), white to cream on Salt Sabouraud dextrose agar upon 7 days of incubation at 40°C (C) (5).
3.3. DNA Extraction
The DNA of the yeast isolates was extracted by the boiling technique. Then, several procedures, including the suspension of a new yeast colony in 50 μL distilled water, boiling at 95°C for 20 min, and centrifugation at 5000 rpm for 5 min, were conducted. The supernatant was stored at -20°C and used as the DNA template (25).
3.4. C. auris-specific PCR
All extracted DNAs from yeast isolates were subjected to amplification by direct C. auris-specific PCR by the primer pairs, namely F250 (5’-ATTTTGCATACACACTGATTTGG-3’) and R250 (5’-AATCTTCGCGGTGGCGTT-3’). Regarding the PCR temperature program, the initial denaturation was performed at 95°C for 5 min. Then, 35 cycles of 94°C for 15 sec and 60°C for 30 sec was done. In the next stage, elongation were carried out at 72°C for 30 sec, as well as a final extension step at 72°C for 5 min. All the products of PCR were electrophoresed on 1% agarose gel stained with 0.5 µg/mL ethidium bromide (5). Then, the internal transcribed spacer PCR-RFLP was used to identify the yeasts isolated on CHROM agarTM Candida medium with the restriction enzymes MspI (Figure 2) (26).
4. Results
In this study, 32 immunocompromised patients were screened for skin surface colonization, who were evaluated by culture and C. auris-specific PCR. The average age of the samples was eight months to 71 years. The majority of the patients were under the age of 10 years. There were 10 male cases, 11 female cases, and 11 children. Only cases with underlying disease, including (patients with weak immune systems or immunodeficiency, were studied. In addition, 15 of 32 patients had valid results by culture, and patients with negative culture were excluded. The skin swabs of patients in the groin (13 swabs), axilla (4 swabs), nare (1 swab), and tongue (1 swab) were positive based on culture assay (SDA). Fungal species present were determined on skin surfaces. The results showed that the most common fungal types contaminated on skin surfaces were Candida albicans (n = 11), Candida glabrata (n = 4), mix (C. albicans and C. glabrata: n = 2), mix (C. albicans and C. parapsilosis: n = 1), and mix (C. albicans and C. glabrata and C. tropicalis and C. parapsilosis: n = 1), but C. auris was not isolated (Table 2).
| Source / No. of Samples | No. of Positive Culture (yeast) | Causative Organism / Frequency |
|---|---|---|
| Groin swabs (n = 31) | 13 swabs | Candida albicans (n = 8) |
| Candida glabrata (n = 2) | ||
| Mix (C. albicans & C. glabrata) (n = 2) | ||
| Mix (C. albicans & C. glabrata & C. tropicalis & C. parapsilosis) (n =1) | ||
| Axillary swabs (n = 31) | 4 swabs | Candida glabrata (n = 2) |
| Candida albicans (n =1) | ||
| Mix (C. albicans & C. parapsilosis) (n =1) | ||
| Nare swabs (n = 2) | 1 swabs | Candida albicans (n =1) |
| Tongue swab (n = 1) | 1 swabs | Candida albicans (n =1) |
| Ear swab (n = 1) | 0 | 0 |
The results indicated that the colonized patients had many live C. albicans on their skin (axilla/groin). The dominant pathogen in inpatient clinical specimens was markedly C. albicans with C. glabrata, C. parapsilosis, and C. tropicalis. C. auris is not a common cause of systemic or superficial fungal infections in Iran. Most C. albicans isolated from the skin swabs were obtained through recovery from the groin, axillary, and other body. In 13 out of 15 subjects, gynecological colonization occurred more frequently. The findings showed the groin as the preferred site of C. albicans colonization compared to the axilla in the colonized subjects (representing moist areas).
Interestingly, only one patient had a positive result for at least one site (one groin) of the 66 swab samples cultured in Salt SDA (S.N.M) (at 40°C). A positive result was identified for C. glabrata when the RFLP-PCR assay was repeated using the stored DNA specimen for this patient. This feature indicates the high ability of this species to tolerate salinity and high temperatures compared to C. auris, which grows significantly in Salt Sabouraud with D-Mannitol as a carbon source.
5. Discussion
C. auris is recognized as a colonizing organism and a source of infection in people in healthcare systems worldwide (2). These infections were linked to invasive medical devices, mechanical ventilation, extended stays in ICUs, and prior exposure to broad-spectrum antibiotics (27). Furthermore, the COVID-19 pandemic has provided excellent conditions for C. auris to propagate (8). Given the limited capacity of ICUs for the number of COVID-19 patients in need of critical care and the difficulty in implementing conventional infection prevention and control measures, it is possible to unintentionally facilitate the silent spread of C. auris through the lengthy utilization of personal protective equipment by healthcare staff (13).
In this study, a C. auris-specific PCR assay and salinity-rich culture were used to effectively suppress other microorganisms and isolate C. auris, using a combination of high temperature and salinity. Using the culture-based method and specific-species PCR, no C. auris strains were isolated from immunocompromised patients admitted to the ICU. Although nobody can guaranty the C. auris as a non-fungal infectious agent forever, itis not a common cause among patients with systemic or superficial fungal infections in Iran.
As mentioned, C. glabrata was the only species growing poorly in Salt SDA with D-Mannitol as the carbon source at high temperatures and in the salinity medium. These findings are consistent with those of Welsh et al. (24) confirming salinity-rich culture processing of skin swabs does not eliminate the development of species other than C. auris.
The results revealed that the patients’ colonized skin (axilla/groin) and mucosal (nares) surfaces had many live C. albicans cells. Carriage or infections with these isolates may lead to compromised treatment choices, and high death rates among cases (5 - 10% of known colonized patients develop invasive infections) (28). C. albicans mainly cause invasive candidiasis and affects the most vulnerable cases in health care settings (eg, the patients in critical condition). Patients with infection or colonization should be detected early to manage with a weak immune system and prevent transmission of environmental contamination from patient to patient (29).
Further research is required to fully comprehend the mechanism of colonization. Infection prevention in healthcare settings should be directed by rapid detection because C. auris is resistant to various antifungal drugs and is also challenging to treat. Laboratories should use proper procedures to determine all cultured Candida species recovered from other body parts to early detect C. auris outbreaks. This study was limited by the small number of samples collected, as it was impractical to visit patients regularly and collect samples due to COVID-19 global prevalence and the hospital quarantine conditions.
5.1. Conclusions
According to the results, C. auris-specific PCR was a successful fast substitute for culturing to identify C. auris in the clinical skin swabs of the axilla/groin. It is possible to obtain results by C. auris-specific PCR in less than 2.5 h, with a remarkable improvement compared to culturing with the 14 days needed.