Dear Editor,
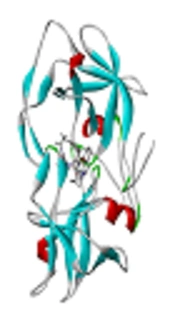
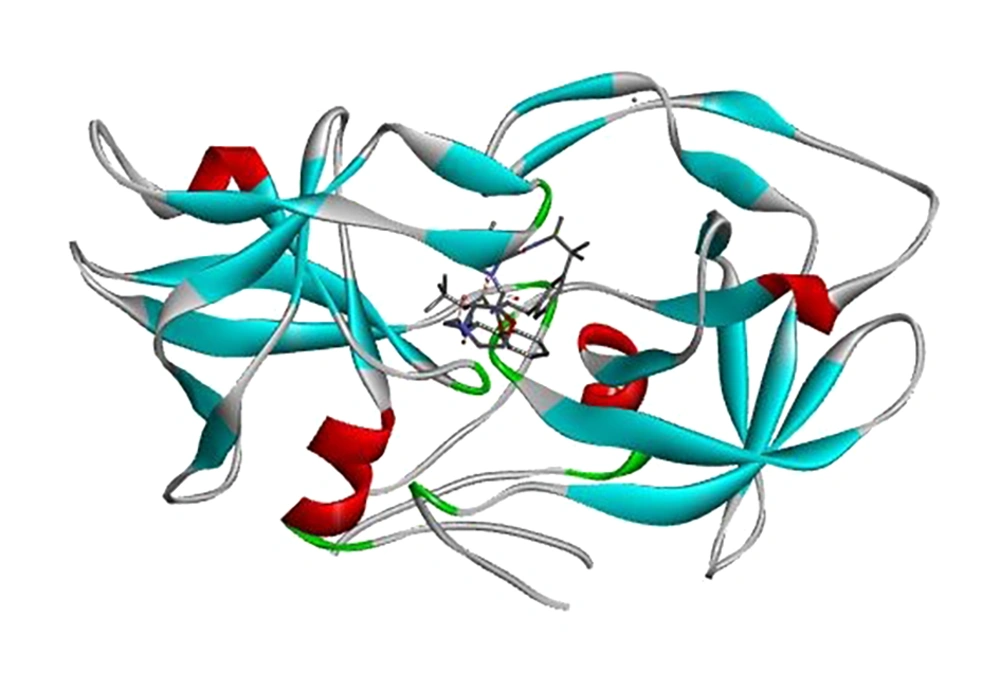
As you know, human T-lymphotropic virus type 1 (HTLV-1), as a type C retrovirus was first isolated from patients involved with cutaneous T-cell lymphoma in 1980 (1). From among all of infected individuals by this virus, approximately 90% remain asymptomatic; some 5% developed to acute status, adult T cell leukaemia/lymphoma (ATLL), and about 4% developed to chronic status, as named HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP), which causes a viral progressive paralysis. Further, about 1% of individuals are afflicted to uveitis (HU) and other diseases with unestablished associations such as arthropathy, pneumopathy, dermatitis, exocrinopathy and myositis (2, 3). HTLV-1 protease (PR) as a homodimer aspartic protease, has a pivotal role for maturation of infected virus. Nowadays, designing of protease inhibitor composition, has been focused on the blocking of protease activity (4).
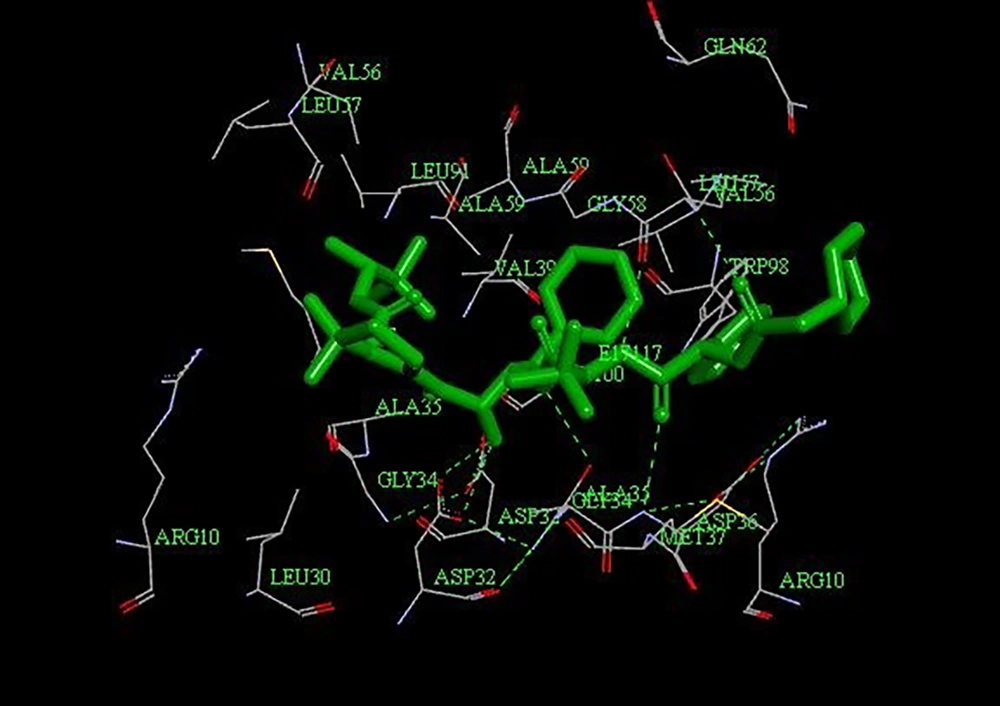
Briefly, three dimensional structure of HTLV-1 protease with accession number 3WSJ, received from Protein Data Bank (PDB). By use of Discovery studio software, H2O molecules and ligand were deleted. Eventually, remaining protein was selected as our interest receptor. Then, five groups of antiviral protease drugs, were evaluated; these are including anti-protease HIV and HCV virus, peptidomimetic inhibitors, Kni-10729 and Kni-10683 inhibitors, as well as their derivatives, modified Kni-10729 and Kni-10683 inhibitors (Table 1). Using of ChemDraw 8.0 software and based on protease active site (consist of seven amino acids, leucine 30, alanine 35, valine 39, tryptophan 98, glycine 34, aspartic acid 32, aspartic acid 36 in the flap region) Peptidomimetic inhibitors were designed, so that, they contain just arginine and histidine residues; highly positive charge of these peptides is necessary for penetration into the cell. In next step, by Hyperchem software, Polack Ribiere algorithm and MM+ (RMS = 0.05 kcal mol-1) model, peptidomimetic, and also, inhibitors based on Kni-10729 and Kni-10683 molecules, were optimized. In order to increasing of anti-viral properties, Fluor- hydroxyl groups was added to their structure. As a necessity for production of synthetic compounds, physicochemical characteristics, specificity, solubility, stability, bioavailability and non-toxicity for humans, was done through online servers, SwissADME, Violation and Lazer. Finally, via Autodock vina software, and an angstrom network with coordinates of X = 34.528, Y = 4.472 and Z = 26.278 with 22 × 42 × 22 was done. Docking was done based on Lamarckian genetic algorithm Parameters, and binding free energy (ΔGb) was determined, ΔGb = 2.3 RtlogKd (5).
| Name | ∆Gb | References |
|---|---|---|
| Indinavir | -11.3 | (4, 6) |
| Quinavir | -8.7 | Swiss similarity (FDA approved) |
| Boceprevir | -7.9 | (5) |
| Grazoprevir | -11.3 | (5) |
| Statine based inhibitor | -7.9 | (7) |
| Allophenylnorstatine | -9.5 | (7) |
| Kni-10729 | -9.5 | (7) |
| Kni-10683 | -9.9 | (7) |
| Arg-Arg | -7.5 | Current study |
| Arg-Arg:F | -5.8 | Current study |
| Arg-His-Arg | -7.2 | Current study |
| Arg-His-Arg:F | -6.8 | Current study |
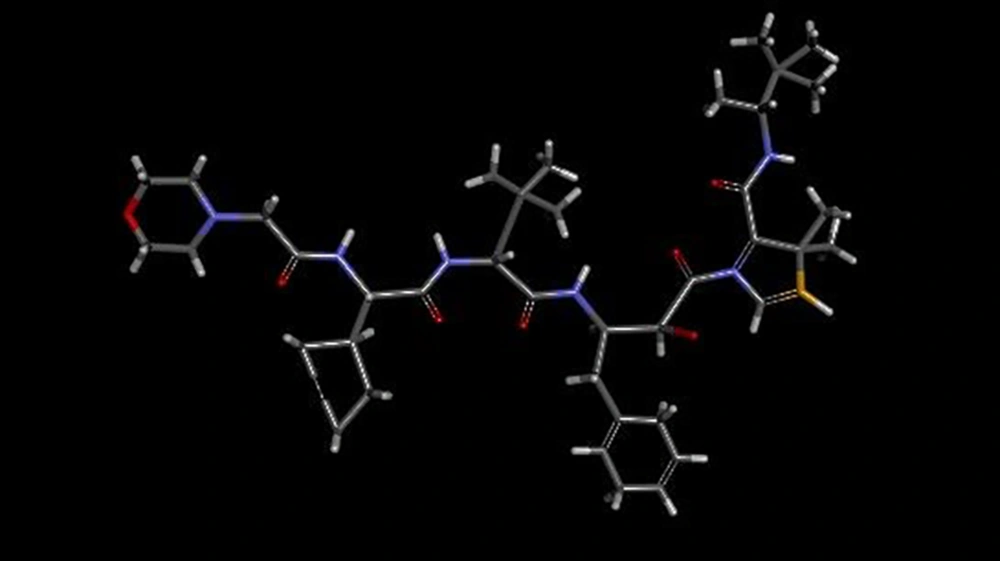
| Modified Kni-10729 | -12 | Current study (Figure 1) |
| Modified Kni-10683 | -11.2 | Current study |
According to related studies, HTLV-1 protease is a homodimer protein, each chain (A and B) consisting of 125 residues. Synthetic molecules that occupy the active site of enzyme, can be considered as HTLV-1 anti-protease drugs (6). In the present study, affinity of 14 different compounds was evaluated for occupation of active site of enzyme; based on results, Kni-10729-derived inhibitor (-12) (Figure 2) and Peptidomimetic Arg-Arg:F (-5.8) have the highest and lowest affinity with protease active site respectively.
Three-dimensional structures of HTLV-1 and HIV proteases, are relatively (approximately 38%) similar to each other. Therefore, it was expected that, HIV anti-protease drugs could be inhibited HTLV-1 protease, which this hypothesis has been proven in in-vivo experiments, too (6, 7). In our study, the binding affinity ratio of Indinavir (as FDA approved drug) to active site of HTLV-1 protease was -11.3 kcal mol-1. Nonetheless, Peptidomimetic inhibitors had least binding affinity ratio to HTLV-1 protease (Table 1). Collectively, the results showed that, Indinavir, Grazoprevir, as well as modified Kni-10729, had the highest binding affinity ratio to amino acids (especially Gly 34, Asp 32, and Asp 36) in protease active site, compared to other ligands (Figure 3). Unfortunately, due to the lack of proprietary drugs against HTLV-1, patients use anti-HIV drugs, which are not fully effective on HTLV-1 infection. The main goal of this study was to compare different anti-proteases against HTLV-1. With regard to present results, modified Kni and HIV-1 and HCV inhibitors, possessed highest affinity to binding and inhibition of active site of HTLV-1 protease. As well, with attention to structural differences between proteases of them, it is more need studies for designing of effective drugs against HTLV-1 protease.