1. Background
Colorectal cancer (CRC) has been recorded as one of the global top 10 cancers allocating rank four after skin, lung, and breast (1, 2). In 2017, 1.8 million incident cases of CRC along with 896000 deaths were reported. CRC is more prevalent in men than women (1/26 for men vs. 1/40 for women) (3, 4). Family history of the disease, inflammatory bowel disease, obesity, and alcohol and tobacco consumption are well-documented risk factors of CRC (5-7). Although this type of cancer has mostly been observed in adults, CRC has been diagnosed at any age (8). Dysregulation of genes may affect the cellular behavior and promote tumorigenesis (9, 10). Long noncoding RNAs (lncRNAs) are lengthy non-protein coding transcripts which are actively involved in modulating critical cellular signaling pathways containing cell cycle, apoptosis, and differentiation (11, 12). Abnormal expression levels of lncRNAs have been described in different human tumors (13). HOX transcript antisense RNA (HOTAIR) is one of the well-studied lncRNAs which has been dysregulated in a variety of solid tumors (13, 14). The literature demonstrated that HOTAIR is associated with apoptosis. Increased expression of HOTAIR may suppress apoptosis (15, 16)]. Gene silencing through RNA interference (RNAi) strategy is recently suggested as the therapeutic method against cancer (17, 18). This is a non-invasive approach which can effectively target the critical pathways of cell proliferation, growth, or invasion (19).
2. Objectives
The present study aimed to investigate the effect of HOTAIR silencing in triggering apoptosis through targeting Bax and Bcl2 in cellular model of CRC.
3. Methods
3.1. Cell Lines/Culture Condition
The cellular model of CRC Caco-2 was purchased from the national cell bank of the Pasteur Institute, Tehran, Iranian Biological Resource Center. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Life Technologies, USA) containing 2mM Glutamine, 10% FBS, 1% penicillin, and 1% streptomycin (Life Technologies, USA). Cells were incubated at 37°C and atmosphere of 5% CO2.
3.2. HOTAIR siRNA Transfection
The 3 × 104 cells were plated onto the 6-well plates and treated with 50 nM siRNA duplexes (siHOTAIR I, SASI Hs02 00380445) for 48h using LipofectamineTM 2000 (Invitrogen, USA) according to the user guideline. Concentration of siRNA was set based on the concentration in our pilot research, and found this concentration more suitable for knocking out.MISSION® siRNA Universal Negative Control #1, SIGMA/SIC001) was used as negative control (20). The mock test was defined as cells treated with lipofectamine and without siRNA.
3.3. RNA Extraction and Real-Time Polymerase Chain Reaction
Following 48h of treatment with siRNA, total RNA was extracted from transfected cells using Trizol reagent (Invitrogen, USA) based on manufacturer protocol. The RNAs were reverse transcribed to cDNA through Primer-Script one step real-time polymerase chain reaction (RT-PCR) kit (Biofact). The cDNA samples were amplified using the SYBR green kit (Biofact) during real-time PCR. The primers used in this study are listed in Table 1. Relative gene expression was calculated as 2−ΔΔCt.
| Target Gene | Sequence Primer | Amplicon Length |
|---|---|---|
| HOTAIR | F':AGGCCCTGCCTTCTGCCT | 147 bp |
| R:TGCTCTCTTACCCCCACGGA | ||
| BAX | F':TTCATCCAGGATCGAGCAGG | 103 bp |
| R:TGAGACACTCGCTCAGCTTC | ||
| BCL2 | F:GAGGATTGTGGCCTTCTTTG | 143 bp |
| R:CGTTATCCTGGATCCAGGTG | ||
| GAPDH | F:GTGAACCATGAGAAGTATGACAAC | 123bp |
| R:CATGAGTCCTTCCACGATACC |
3.4. Statistical Analyses
Data were analyzed using a one-way analysis of variance with Newman-Keuls correction for multiple comparisons using the GraphPad Prism version 6 (GraphPad Software, USA). The nonparametric tests were used if normal distribution was not met. P-value less than 0.05 was considered as a statistically significant difference.
4. Results
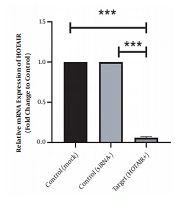
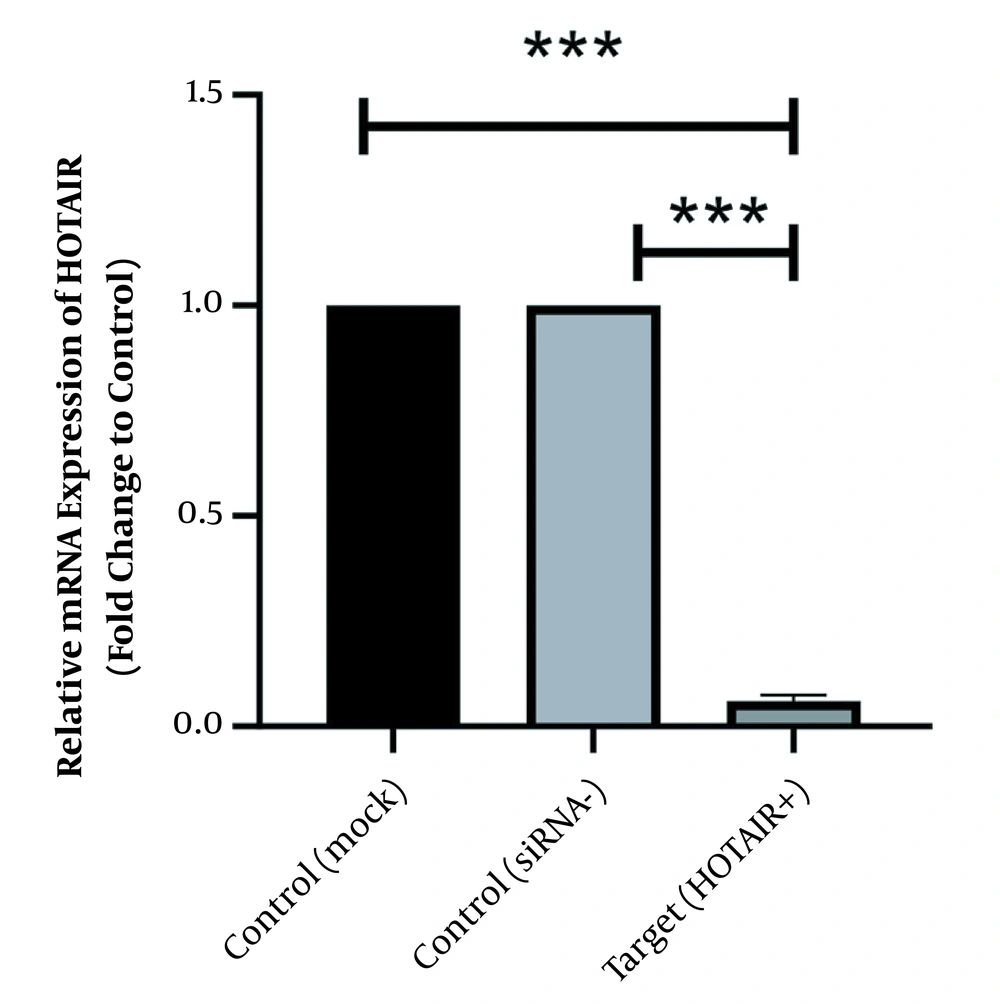
As illustrated in Figure 1 HOTAIR is strongly expressed in normal culture of Caco-2 cells before siRNA treatment. However, its expression level has been significantly downregulated since the HOTAIR-specific siRNA was added into the culture medium. The fold-change was 0.06 ± 0.011 (P-value < 0.001).
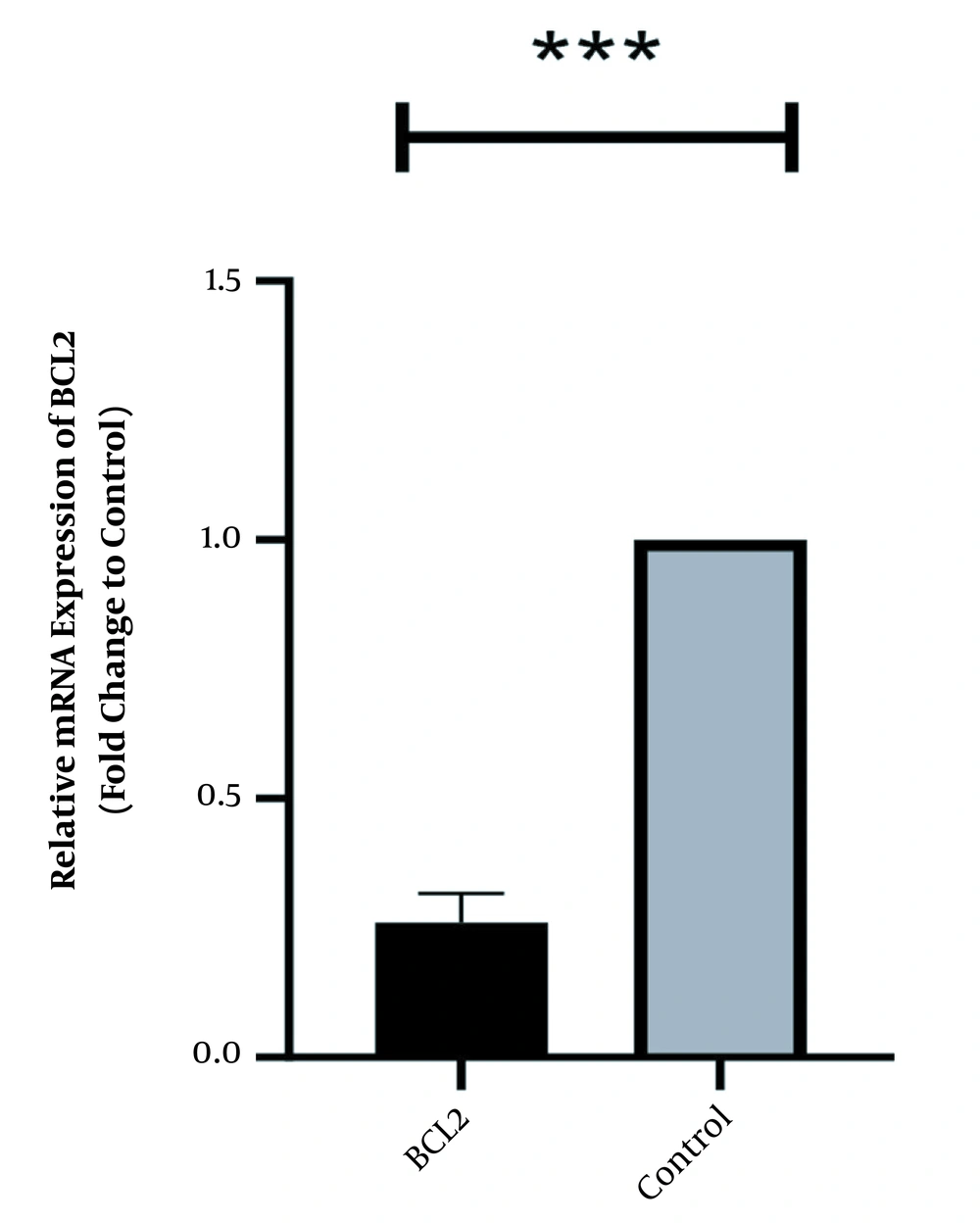
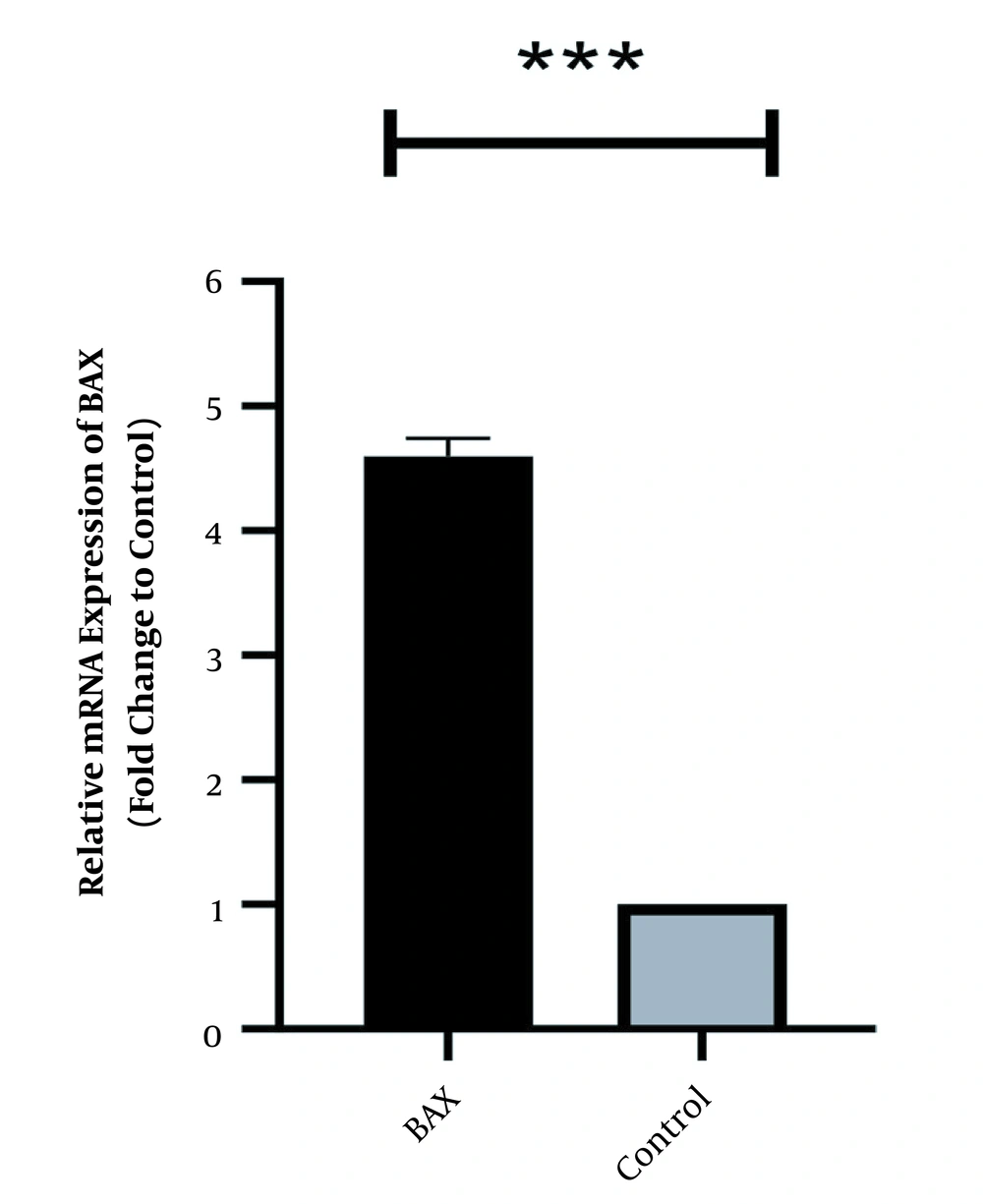
In the same way, the expression level of Bcl2 was high in normal culture of Caco-2 and downregulated after 48 h of treatment with HOTAIR-specific siRNA. The corresponding fold-change was 0.26 ± 0.042 (P-value < 0.001) (Figure 2). Conversely, the expression level of Bax was weak in Caco-2 cells and significantly increased as the cells were treated with HOTAIR-siRNA. The calculated fold-change was 4.6 ± 0.031 (P-value < 0.001) (Figure 3).
5. Discussion
The present study showed the effects of HOTAIR silencing on the induction of apoptosis. Two well-known modulators of apoptosis, Bax, and Bcl2, were traced. Data showed Increased expression of Bax and decreased expression of Bcl2. Previously studies indicated that inhibition of HOTAIR can promotes apoptosis through different signaling pathways. However, the underlying mechanism is not well-understood in CRC. The Bax protein, which is a pro-apoptosis factor, activates apoptosis, while the Bcl2 is an anti-apoptotic protein. The Bax/Bcl2 ratio acts as a controller which decides whether cells turn on apoptosis or not. At the lower levels of ratio, the apoptosis is suppressed (21). Similarly, previous studies showed the high expression of Bcl2 in different cancers, which made them resistant to chemo- or radio-therapy (22). This is the reason that the oncoprotein Bcl2 has been proposed as therapeutic target for cancer through apoptosis induction (23). We also found that Bax had low level of expression, and its expression increased around 4-fold following siRNA treatment. The low expression of this gene was previously reported in colorectal and lung cancers (24, 25). It has been demonstrated that Bax permeabilizes the mitochondrial membrane to release cytochrome c and eventually apoptosis (26). Therefore, agents that activate Bax can be considered as promising anticancer drug. Several drugs have been confirmed or candidate to activate Bax directly or indirectly. Bortezomib, Ixabepilone, and Sorafenib are three examples of Bax activators (26). In conclusion, high level of Bcl2 protein and low level of Bax can inhibit apoptosis in cancer cells, while low amount of Bcl2 and high amount of Bax make cancer cells susceptible to the apoptosis. As we observed here, HOTAIR silencing can act as apoptosis inducer, which is partly due to the activation of Bax and inhibition of Bcl2. It would be ideal to consider the growth inhibitory effect of HOTAIR silencing on mouse models for CRC. Such a study might clarify whether HOTAIR silencing can provide a promising strategy to cure cancers. It is necessary to remind that apoptosis is only one among hundreds of molecular pathways under the control of HOTAIR. This lengthy noncoding transcript regulates variety of cellular pathways, including cell cycle, differentiation, and growth (27). Therefore, HOTAIR silencing may inhibit the cancer progression through different pathways, which can be considered in future studies.