1. Background
Prenatal genetic diagnosis has recently been issued in obstetric practice. Most prenatal diagnostic programs have typically been focused on conditions such as fetal chromosomal aneuploidies and monogenic diseases of high prevalence in various populations (1, 2). The existence of cell-free fetal DNA (cffDNA) in maternal circulation was discovered by Lo et al. about 20 years ago, and has become a useful molecular material for prenatal genetic diagnosis especially for fetal sex determination (3), the RhD status of fetuses, screening of pregnancy-related complications, fetal diseases like trisomy 21, sex-linked disorders and single gene disorders (2, 4-8). Non-invasive declaration of fetal sex at early gestational ages could help eliminate the need for invasive methods (e.g. molecular testing of chorionic villi at the 11th week of gestation) (9), manage congenital adrenal hyperplasia where female fetuses could be treated by corticosteroids at the 6th week of gestation, and assess ambiguous genitalia identified by ultrasound examination (10, 11). As an example, carrier mothers of serious X-linked conditions such as Duchenne muscular dystrophy require genetic prenatal diagnosis only when the fetus is male (2, 12). Furthermore, CffDNA can be detected as early as the fourth week of gestation and its concentration increases throughout pregnancy until two hours after delivery in a time for proper clearance from maternal plasma (13, 14). The concentration of cffDNA, which contains 10% of the DNA in maternal plasma is much more than that in the cellular fraction of maternal blood (4, 15). That is why most investigators have focused on cffDNA rather than fetal cells. Fetal gender has been determined using PCR amplification of fetal-specific Y-derived sequences like SRY and DYS14 with sensitivity and specificity of almost more than 95%. According to all related studies conducted so far, pregnancies with no chromosome Y DNA have been assumed to be bearing female fetuses (16-19). Originating from trophoblasts, the cffDNA in maternal blood consists of short fragments typically fewer than 200 bp in length (20).
2. Objectives
We preferred to use markers short enough to be amplified by PCR with high probability. Accordingly, ten X chromosome mini-STRs and nine Y chromosome mini-STRs and three non-STR markers (SRY, DYS14 and amelogenin) were selected to trace fetal DNA in maternal plasma after assessing their size, variability and distribution throughout the X and Y chromosomes. Algorithm-based approach of mini-STR genotyping during the present study was designed to cut down on false negative and or positive reports of fetal gender determination.
3. Patients and Methods
3.1. Blood Collection
Peripheral blood samples were obtained from 106 pregnant women (during the period between the third and fifteenth week of gestational age) and their husbands who had referred to the obstetric clinics of Hafez hospital(Shiraz, Iran). All singleton pregnant women who had no abortion, transplantation, and blood transfusion were included in our study. Validated by medical ethic committee of the university (research proposal No. 90-5531 approved in 2012. 02. 10), informed written consent was obtained from each person participating in this study before blood sampling. We collected each blood sample in a completely sterile tube containing Ethylenediaminetetraacetic acid (EDTA), and transferred the samples to the laboratory.
Plasma was separated from the pregnant women’s whole blood samples as soon as possible by centrifugation in two successive steps (18000 g for 10 minutes [optional: storing at -80°C] and 2700 g for 45 minutes). The buffy coat of pregnant women (after plasma removal) and also their husband’s whole blood samples were prepared for DNA extraction or stored at -20°C for subsequent use.
3.2. DNA Extraction
DNA was extracted from 1 mL plasma samples using QIAamp DNA blood mini kit (QIAGEN, Hilden, Germany) according to the blood and body fluid protocol with minor modifications. The final volume of the eluted DNA was about 50 μL. Using the manual salting-out method, we extracted DNA of the buffy coat and husbands’ whole blood samples. The DNA quantity and quality was measured by Nano-Drop (ND1000; NanoDrop technologies, Wilmington, DE) and proved to be applicable for PCR analysis. To avoid DNA contamination, all procedures were performed almost exclusively and consistently in an isolated partition by female staff members with strict precautions.
3.3. Mini-Short Tandem Repeats, Primers and Polymerase Chain Reaction
We employed ten X-chromosome mini-STRs and 12 Y-chromosome markers (nine mini-STRs and three non-STRs) for fetal DNA detection in maternal plasma. Furthermore, X and Y mini-STRs were opted amongst various forensic and related studies due to their applicability in fragmented DNA discovery.
Incidentally, using specific primers (Tables 1 and 2) which were designed and synthesized in a way that would narrow the mini-STR amplicon size as short as possible (less than about 160 bp in length) to increase the detectability of fragmented fetal DNA in maternal plasma. PCR was performed with two different thermal conditions. The PCR for amplifying X mini-STRs was implemented as follows: initial heating at 95°C for 5 minutes, 30 cycles of 95°C for 1 minute, 60°C for 30 seconds, and 72°C for 20 seconds and a final extension at 60°C for 40 minutes. For amplifying the Y mini-STRs and non-STR markers, the conditions were 95°C for 5 minutes for an initial denaturation, 30 cycles of 95°C for 1 minute, 59°C for 30 seconds, and 72°C for 20 seconds and a final extension at 60°C for 40 minutes. Each reaction (25 μL) for amplifying either X mini-STRs or Y markers consisted of: 2.5 μL of 10X PCR buffer, 1.5 mM of MgCl2, 0.3 mM of dNTPs, 2 U of Smart Taq polymerase, 0.3mM of each primer, and 4 μL of plasma DNA (about 24 ng) or 2.5 μL of each parent’s DNA samples. Afterward, we used Polyacrylamide gel electrophoresis (PAGE) for size separation of PCR products (21).
| Classification of Mini-STR | Primer Sequences (5' - 3') | Motif Repeat | Product Size, bp |
|---|---|---|---|
| Y Mini-STRs | |||
| DYS389b | (TCTG) 5 (TCTA) 12 | 122 | |
| F | CCAACTCTCATCTGTATTATCTATG | ||
| R | TATTATACCTACTTCTGTATCCA | ||
| DYS459 | (ATTT) n | 136 - 156 | |
| F | CAGGTGAACTGGGGTAAATAAT | ||
| R | TTGAGCAACAGAGCAAGACTTA | ||
| DYS446 | (TCTCT) n | 85 - 160 | |
| F | TATTTTCAGTCTTGTCCTGTC | ||
| R | GAGACTCTGTCTGAAGAGAG | ||
| DYS426 | (GTT) n | 85 - 110 | |
| F | GGTGACAAGACGAGACTTTGTGT | ||
| R | CTCAAAGTATGAAAGCATGACCA | ||
| DYS438 | (TTTTC) n | 95 - 140 | |
| F | TGGGGAATAGTTGAACGGTAA | ||
| R | GGCAACAAGAGTGAAACTCCA | ||
| DYS481 | (CTT) n | 115 - 158 | |
| F | AGGAATGTGGCTAACGCTGT | ||
| R | ACAGCTCACCAGAAGGTTGC | ||
| DYS505 | N-7-(T)-[TCCT] n-2-N24 | 97 - 125 | |
| F | CTCTGTTCTTTTTCTCTCCTTCC | ||
| R | AGGTTCGAGTCAGTTCACCA | ||
| DYS441 | N7-[TTCC] n-2-(T)-N-7 | 91 - 119 | |
| F | CAAATTCTCAGGCATTGCAG | ||
| R | GGGAGAGAAGGAGGAAGGA | ||
| DYS392 | (TAT) n | 94 - 130 | |
| F | AAAAGCCAAGAAGGAAAACAAA | ||
| R | GAAACCTACCAATCCCATTCCTT | ||
| Non-STR Markers | |||
| SRY | Non-repeat sequences | 76 | |
| F | TCCTCAAAAGAAACCGTGCAT | ||
| R | AGATTAATGGTTGCTAAGGACTGGAT | ||
| DYS14 | Non-repeat sequences | 84 | |
| F | GGGCCAATGTTGTATCCTTCTC | ||
| R | GCCCATCGGTCACTTACACTTC | ||
| X and Y Marker | |||
| Amelogenin | Non-repeat sequences | 106, 112 | |
| F | CCCTGGGCTCTGTAAAGAATAGTG | ||
| R | ATCAGAGCTTAAACTGGGAAGCTG | ||
Primer Sequences, Y-Marker Allele Range and Length of Polymerase Chain Reaction Product Size
| Classification of X Mini-STRs | Primer Sequences (5’ – 3’) | Motif Repeat | Product Size, bp |
|---|---|---|---|
| DXS7133 | TAGA | 76 - 100 | |
| F | AGCTTCCTTAGATGGCATTCA | ||
| R | GTTTTTAACGGTGTTCATGCTT | ||
| DXS8378 | CTAT | 95 - 11 | |
| F | GCTCCTGGCAGGTCACTATC | ||
| R | GCGACAAGAGCGAAACTCCA | ||
| DXS7132 | (TCTA) x-(TCA) (0-l)-(TCTA) 2 | 135 - 163 | |
| F | CCTCCTTAATAGTGTGAGCCCAT | ||
| R | GTCAACGTTCTCCAGAGAAACAGA | ||
| DXS7423 | CCAT | 95 - 115 | |
| F | AGATTTCCTCCCCATCCATC | ||
| R | GTTGTCACACAAATAAATGAATGAGT | ||
| GATA31E08 | (AGGG) x-(AGAT) y | 95 - 131 | |
| F | CAGAGCTGGTGATGATAGATGA | ||
| R | CTCACTTTTATGTGTGTATGTATCTCC | ||
| DXS6789 | (TATC)(0-l)-(TATG)x-(TATC)y | 124 - 168 | |
| F | CCTCGTGATCATGTAAGTTGG | ||
| R | CAGAACCAATAGGAGATAGATGGT | ||
| GATAD05 | TAGA | 122 - 150 | |
| F | TAGTGGTGATGGTTGCACAG | ||
| R | ATAATTGAAAGCCCGGATTC | ||
| DXS6803 | (TCTA) x-(TCA) (0-l)-TCTA | 102 - 30 | |
| F | GAAATGTGCTTTGACAGGAA | ||
| R | CAAAAAGGAACATATGCTACTT | ||
| DXS101 | (CTT) x-(ATT) y | 126 - 177 | |
| F | TCTCCCTTCAAAAACAAAGATAA | ||
| R | TGCATATTCTGCGCATGT | ||
| GATA165B12 | AGAT | 90 - 110 | |
| F | TCATCAATCATCTATCCGTATATCA | ||
| R | GAAGTTGACTGTGATTCCTGGTTT |
Primer Sequences, X-Mini Single Tandem Repeat Allele Range and Length of Polymerase Chain Reaction Product Size
3.4. Polyacrylamide Gel Electrophoresis
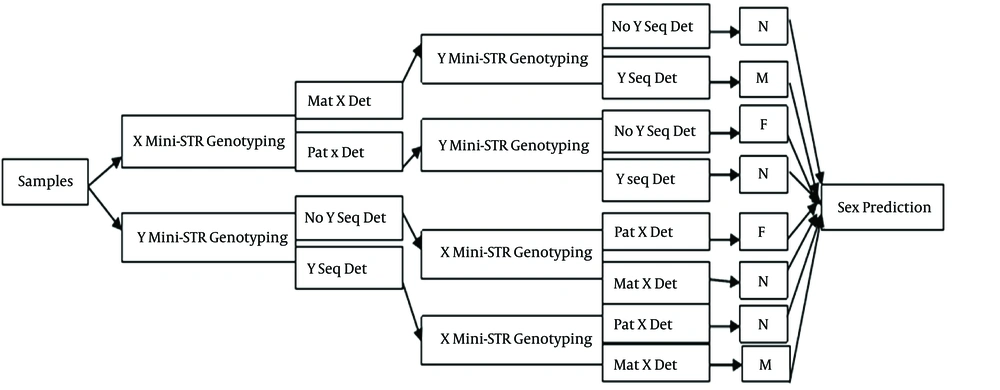
When 10% polyacrylamide gel was prepared, 7 μL of the PCR product of each mini-STR was injected into its own well, which had been configured on vertical electrophoresis apparatus (PROTEAN II Xi cell; Bio-Rad Laboratories, Hercules, CA), and with a constant current of 130 mA, the PCR products started to move downwards through acrylamide gel pores. The gel was then stained with the silver staining method. Finally, the genotypes and sex of each fetus were determined by comparison with his or her parents’ pattern of mini-STR alleles (supplemental data Figure 1 A and B) and according to the algorithm (named X-Y algorithm) chart illustrated below (Figure 1). This algorithm was designed in five steps. Step 1, Input material (sample); step 2, X (or Y) mini-STR genotyping (if Y markers are genotyped in this step the next step would be X-markers genotyping); Step 3, Y (or X) mini-STR genotyping, following and confirming step 2 results; Step 4, comparing step 2 and 3 and gathering data; Step 5, output (sex prediction).
3.5. Statistical Analysis
The SPSS 16.0 (SPSS Inc, Chicago, IL), Stata, SE 12.0 and prism 4 (GraphPad software, San Diego, CA) software were utilized for data analysis. The Fisher exact test (two-sided) was used to determine sensitivity and specificity, positive predictive value (PPV) and negative predictive value (NPV) of the method with 95% confidence intervals (CIs). Descriptive statistics were applied for each marker. In addition, sensitivity, specificity and informativeness of each marker were also calculated.
4. Results
Birth outcome showed that amongst all samples, 46.2% were carrying male fetuses, 5.6% had a miscarriage (excluded from the statistical analysis), and the rest (48.1%) were carrying female fetuses.
4.1. Mini-Single Tandem Repeat Genotyping Results
Algorithm-based mini-STR genotyping method identified 47 (46.2%) pregnant women with male fetuses and 49 (48.1%) pregnant women with female fetuses. Therefore, the sensitivity and specificity of the modified conventional PCR considering the pregnancy outcome were calculated to be 95.9% and 98%, respectively. Amongst all samples, fetal gender of four cases (at five, five, six, and seven weeks of gestation) failed to be determined, and in one case (at nine weeks of gestation) fetal sex was incorrectly conjectured.
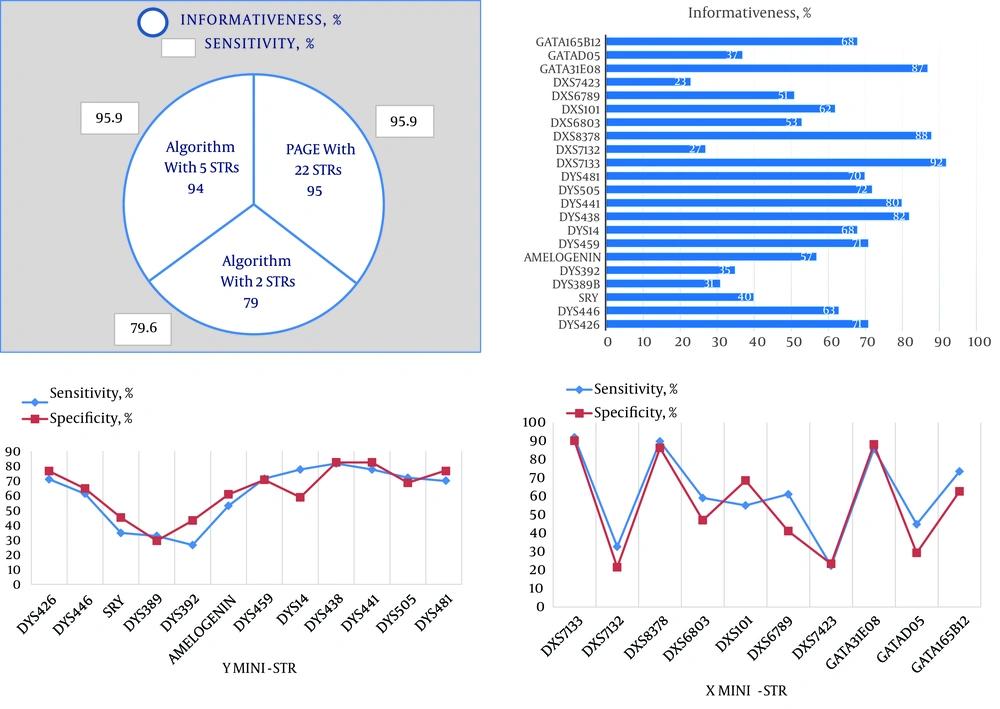
To show applicability of our X-Y algorithm and to obtain minimum number of markers in a test panel with maximum sensitivity and specificity for fetal sex prediction, we computed the sensitivity of the algorithm and cumulative informativeness of markers and other parameters (Figure 2, Table 3). In this approach, we compared two possible ways of using markers in X-Y algorithm by which we could achieve a reasonable and statistically significant number of markers. One supposed condition included one X mini-STR and one Y mini-STR (least amount of markers suited for the algorithm) while in the other supposed condition significant amounts of markers (five markers: three X mini-STRs and two Y mini-STRs) were used in the analysis. If we used five instead of twenty-two markers we would be able to predict sex with no significant difference in algorithm sensitivity or cumulative informativeness (P = 0.003) in contrast with using two markers (P > 0.05).
A, Comparing sensitivity and informativeness of the algorithm in three conditions and applicability of the X-Y algorithm (by five markers) for sex prediction in future studies and clinical settings; B, informativeness of X and Y mini-STRs; C, sensitivity and specificity of Y mini-STRs; D, sensitivity and specificity of X mini-STRs.
| Results | Informativeness, CI (95%) | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|---|
| PAGE with 22 STRs | 95 (88.7 - 98.4) | 95.9 | 98 | 94.0 | 97.9 |
| Algorithm with DXS7133 and DYS438 | 79 (69.7 - 86.5) | 79.6 | 78.4 | 78.0 | 80.0 |
| Algorithm with DXS7133, DXS8378, GATA31E08 and DYS438, DYS441 | 94 (87.4 - 97.8) | 95.9 | 92.2 | 92.2 | 95.9 |
Applicability of X-Y Algorithm (by Five Markers) for Sex Prediction in Future Studies and Clinical Settingsa
4.2. Marker Assessments for Being Applied in Clinical Settings
The most informative markers (on the basis of informativeness of higher than 80% and according to applicability of the algorithm) for fetal sex determination were DYS441 and DYS438 among Y mini-STRs and DXS8378, DXS7133 and GATA31E08 among X mini-STRs. This demonstrates that the average number of X mini-STRs that were informative for fetal sexing was more than the average number of Y mini-STRs (Figure 2).
The most sensitive markers were DYS438 (81.6%, CI of 95%), DXS7133 (93.9%, CI of 95%), DXS8378 (89.8%, CI of 95%) and GATA31E08 (85.7%, CI of 95%). Therefore, X mini-STRs were shown to have greater sensitivity in fetal DNA detection rather than Y mini-STRs (see details in Table 4, Figure 2).
| Mini-STRs | Informativeness, CI (95%) | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|---|
| Y mini-STRs | |||||
| DYS426 | 71 (61.1 - 79.6) | 65.3 | 76.5 | 72.7 | 69.6 |
| DYS446 | 63 (52.8 - 72.4) | 61.2 | 64.7 | 62.5 | 63.5 |
| SRY | 40 (30.3 - 50.3) | 34.7 | 45.1 | 37.8 | 41.8 |
| DYS389b | 31 (22.1 - 41.0) | 32.7 | 29.4 | 30.8 | 31.3 |
| DYS392 | 35 (25.7 - 45.2) | 26.5 | 43.1 | 31.0 | 37.9 |
| Amelogenin | 57 (46.7 - 66.9) | 53.1 | 60.8 | 56.5 | 57.4 |
| DYS459 | 71 (61.1 - 79.6) | 71.4 | 70.6 | 70.0 | 72.0 |
| DYS14 | 68 (57.9 - 77.0) | 77.6 | 58.8 | 64.4 | 73.2 |
| DYS438 | 82 (73.5 - 89.0) | 81.6 | 82.4 | 81.6 | 82.4 |
| DYS441 | 80 (70.8 - 87.3) | 77.6 | 82.4 | 80.9 | 79.2 |
| DYS505 | 72 (62.1 - 80.5) | 75.5 | 68.6 | 69.8 | 74.5 |
| DYS481 | 70 (60.0 - 78.8) | 63.3 | 76.5 | 72.1 | 68.4 |
| X mini-STRs | |||||
| DXS7133 | 92 (84.8 - 96.5) | 93.9 | 90.2 | 90.2 | 93.9 |
| DXS7132 | 27 (18.6 - 36.8) | 32.7 | 21.6 | 28.6 | 25.0 |
| DXS8378 | 88 (80.0 - 93.6) | 89.8 | 86.3 | 86.3 | 89.8 |
| DXS6803 | 53 (42.8 - 63.1) | 59.2 | 47.1 | 51.8 | 54.5 |
| DXS101 | 62 (51.8 - 71.5) | 55.1 | 68.6 | 62.8 | 61.4 |
| DXS6789 | 51 (40.8 - 61.1) | 61.2 | 41.2 | 50.0 | 52.5 |
| DXS7423 | 23 (15.2 - 32.5) | 22.4 | 23.5 | 22.0 | 24.0 |
| GATA31E08 | 87 (78.8 - 92.9) | 85.7 | 88.2 | 87.5 | 86.5 |
| GATAD05 | 37 (27.6 - 47.2) | 44.9 | 29.4 | 37.9 | 35.7 |
| GATA165B12 | 68 (57.9 - 77.0) | 73.5 | 62.7 | 65.5 | 71.1 |
Informativeness, Sensitivity, Specificity, Positive Predictive Value and Negative Predictive Value of each Y mini-Single Tandem Repeat and X mini-Single Tandem Repeata
Calculating the average number of markers, which were informative for each sample would lead us to ignore unnecessary markers; that is why we analyzed and obtained data shown in Table 5. As implied by Table 5, about seven Y markers (60.66% ± 2.07) and six X markers (58.8% ± 1.98) were the mean minimum number of markers that were informative for predicting fetal gender of each sample (Table 5). Thus, we propose DYS441, DYS438, DYS481, DYS459, DYS505, DYS426, DYS14, DXS8378, DXS7133, GATA31E08, GATA165B12, DXS6789 and DXS101 markers to be employed for fetal sex determination in diagnostic clinics as the first step and then minimum number of markers (according to our studied five markers) can be utilized in a test panel to obtain maximum sensitivity and specificity for fetal sex prediction. It should be mentioned that highly sensitive markers were not ascertained unless we evaluated them along our experiments. Sensitive markers are those that could be easily amplified and detected during each sample gel analysis. Informative markers are those that enabled us to predict fetal genders in comparison with non-informative ones that had no effects on our projections. Therefore, one marker can be a sensitive marker but not necessarily an informative one.
5. Discussion
Results of our previous study confirmed that our modified conventional PCR (mini-STR genotyping) with an algorithmic approach is in some points a more authenticated method when compared to current quantitative real-time (QRT)-PCRs for fetal gender determination and could be engaged in clinical settings (21). This study was designed to verify its practicability for prenatal sex identification of fetuses in diagnostic laboratories with high accuracy and reliability in addition to low cost. In the recent years, fetal sex has been identified by a single non-polymorphic SRY gene (22, 23). Although some clinics still prefer to use SRY for such trials, it is going to be superannuated and replaced by developed methods in which markers such as DYS14, amelogenin, X STR and Y STR are being studied, associatively or alone, using QF-PCR and QRT-PCR to reach more sensitive and specific assays (24-27). Vecchione et al. implemented multiplex QF-PCR, amplifying X-STR (ranged 103 - 250 bp) together with amelogenin gene markers, on 26 pregnant women and considered some markers as informative markers (28). Nair et al. also designed a study to screen the Y specific DYS19, DYS385 and DYS392 STRs to trace male fetus DNA by which they investigated the clearance of fetal DNA from maternal blood after delivery and reported STR sensitivity of more than 91% for each (29). Scheffer et al. combined both PCR results for the Y-linked sequences, SRY and DYS14, for fetal gender determination and concluded that the DYS14 assay targeted a multi-copy sequence and therefore had a higher sensitivity than SRY (30). Deng et al. sought nine Y-STR loci of fetal DNA in maternal blood and found that the numbers of Y-STR loci, where the maximum allelic size was less than 100, 137 and 180 bp, observed in Chinese individuals, were one, five, and nine loci, respectively (31). The present study applied the conventional PCR method specialized for amplifying polymorphic X and Y mini-STRs, and utilized an algorithmic based genotyping approach to find fragmented fetal DNA in maternal plasma with high sensitivity and specificity. Along with other developing studies in this field, the most sensitive and specific mini-STRs and probably for the first time the average needed number of X and Y markers for fetal gender determination were reported in our study; such sensitive markers could collectively be clinically validated candidates in diagnostic processes. The advantages of our study in contrast with up-to-date studies are the shortness of STR amplicons (mini-STRs) and use of both X and Y markers simultaneously in our low-error algorithm for fetal marker genotyping. Consequently, chances of fetal DNA amplification in maternal plasma would increase by means of these mini-STRs and also not detecting Y markers would not directly lead us to the conclusion of existing female fetus in pregnant women. Therefore, in X-Y algorithm no detection of Y STRs in maternal blood has two meanings, firstly we have not been able to detect fetal DNA at all owing to limited or low quantity of fetal DNA in maternal blood and or failure of the extraction and amplification methods, and secondly there have not been any male sequences (Y STRs) detected; instead, identification of paternal X sequences makes us suppose that the fetus is female. According to many studies, early determination of fetal sex is feasible using cffDNA from four weeks, so this technology should be made available to all women at risk of bearing a fetus afflicted to X-linked disorders or metabolic conditions; this eventually reduces invasive procedures by up to 50%. It is also useful to verify genetic sex when there is a suspicion of genital ambiguity on ultrasound. During our study, paternity testing was correctly performed at early gestational ages for a few referred forensic cases (data are not shown). Besides, a few pregnant women with X-linked family history were screened and their fetus gender was correctly determined before chorionic villus sampling or amniocentesis procedures. There was no X aneuploidy sample among our cases; however, using the mentioned X-Y algorithm, abnormal fetuses affected with X-numerical aneuploidies such as turner and Klinefelter (not non-disjunction II cases) might be easily distinguished. It is noteworthy to mention that, sex prediction failures in this study were not irrelevant to extraction materials and methods and also low sensitivity of our conventional PCR and PAGE. Prospectively, we recommend that future studies should be focused on advanced methods for extracting and separating high cffDNA yields and designing an equivalent algorithmic-based QRT-PCR. This will probably solve such method-borne problems and hopefully will change the era of prenatal genetic diagnosis.