1. Background
The West Nile Virus (WNV) belongs to genus Flavivirus of the family Flaviviridae (1). Lineage 1 and 2 of WNV are associated with significant human outbreaks and are designated into at least 5 phylogenetic lineages (2). The genome of WNV is about 11 kb in length with a single-stranded positive-sense RNA, which encodes seven non-structural (NS) proteins and three structural proteins (C, capsid; M, membrane; and E, envelope) (3). The structural proteins constitute the virus particle while the NS proteins are essential for viral replication (4). The WNV is widespread all over the world and is transmitted by birds and mosquitoes (5). More than 34,000 cases have been reported in United States, with an estimated 7 million cases between 1999 and 2016 (6). WNV infection is defined as cases confirmed by laboratories.
On the basis of laboratory diagnosis involving virus identification by reverse transcription PCR (RT-PCR) or specific antibody detection techniques such as enzyme-linked immunosorbent assay (ELISA) or virus-neutralization (7). However, these methods are time consuming and require technical expertise. Hence, a rapid, simple, sensitive, and efficient method is required to reduce the incidence of WNV cases and infection. Loop-mediated isothermal amplification (LAMP) is simple and the most popular isothermal technology for in vitro nucleic acid amplification (8). The LAMP assay is highly sensitive and able to detect low copy numbers of DNA and is also applicable to RNA using reverse transcriptase. Only primers, polymerase, and a heat block are required to perform the LAMP. Advantages of the LAMP method include the ability to amplify nucleic acid with high specificity, efficiency, and rapidity under isothermal conditions (9). In this study, a newly designed primer was used to efficiently diagnose WNV using the real-time reverse transcription loop-mediated isothermal amplification (RT-LAMP) method.
2. Objectives
This study, we found real-time reverse transcription LAMP (RT-LAMP) assay to be more sensitive than conventional RT-PCR, and more rapid and efficient than conventional RT-PCR and real-time RT-PCR. It will be useful for quick detection of WNV infection and will contribute to the epidemiology study.
3. Methods
3.1. Virus Sample and Ethics Statement
The West Nile Virus serum and plasma sample was obtained from Boca Biolistics (Pompano Beach, Florida, USA; https://www.bocabio.com/). Since this study does not involve patients, the use of these samples was reviewed and approved by the Institutional Review Board for the purpose of research. The Catholic Medical Center Office of the Human Research Protection Program (CMC OHRP) of South Korea (approval number MC19SESI0015) supervised all experimental work and sample collection.
3.2. Viral RNA Extraction
A Bio Safety Level 3 (BSL-3) facility was used for in vitro experiments at the Korea Zoonosis Research Institute, Jeonbuk National University. The control RNA viruses were Zika virus, chikungunya virus, influenza virus (H5N1), severe fever with thrombocytopenia syndrome virus (SFTSV), Japanese encephalitis virus (JEV), and dengue viruses 2 and 3 (DENV-2, DENV-3). Viral RNAs were extracted from the viral stocks using QIAamp Viral RNA Mini Kits (#52906; Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
3.3. Design of Primers
A total of 32 genomic sequences of WNV were obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and conserved nucleotide sequence regions were identified using CLC Main Workbench 7 (version 7.6.4.). Six WNV-specific RT-LAMP primers targeting the E gene were designed. These primers consist of two external primers (F3 and B3), two internal primers (FIP and BIP), and two loop primers (LoopF and LoopB) (Table 1). The Korea National Institute of Health (KNIH) standard protocol was used for conventional RT-PCR using the primers WN-21F (5′-TGACAACTTAGTAGGTTTG-3′) and WN-550R (5′-CAGCTGTTGTAATCGTGATG-3′). The real-time RT-PCR primers were as described in previous research (10). The primers NS1-F (5′-GCCATATGGTACATGTGGCTGGGAGC-3′) and NS1-R (5′-GTKATTCTTGTGTCCCAWCCGGCTGTGTCATC-3′) were designed to bind to the conserved region of the NS5 segment of WNV.
| Primers′ Name | Sequence (5′- 3′) | Location in Genome a |
|---|---|---|
| WNV-F3 | AGAATATGGAGAGGTGACAGT | 1500 - 1520 |
| WNV-B3 | AATTCCACAGGAATGGCTC | 1760 - 1778 |
| WNV-FIP | GGAGGTTGAGGTCCATGAACCTGCATACTACGTGATGACTG | 1607 - 1627, 1555 - 1574 |
| WNV-BIP | TGCTGGAAGTACTGTATGGAGGACAATGCTATCACAGACTGCT | 1703 - 1722, 1638 - 1660 |
| WNV_LoopF | ACTCACGATGGACCAAGAAC | 1587 - 1606 |
| WNV_LoopB | GAACCACACGCCACGA | 1687 - 1702 |
aGenome position according to WNV complete sequence (GenBank accession number KY229069).
3.4. PCR and Amplification
Conventional RT-PCR followed the KNIH standard protocol. The reverse transcription was performed at 50°C for 30 min, then 95°C for 15 min. Amplification occurred under 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. For the real-time RT-PCR, the iTaqTM Universal SYBR Green One-Step Kit (#172-5150; BIO-RAD, Hercules, CA, USA,) and a Bio-Rad CFX96TM system were used. The reverse transcription was performed for 10 min at 50°C, and polymerase activation and DNA denaturation for 1 min at 95°C. The amplification followed with 10 s at 95°C for denaturation, and 30 s at 60°C for annealing/extension under 40 cycles. Real-time RT-LAMP was performed for 50 min at 65°C using a WarmStart LAMP Kit (#E1700; New England Biolabs, Ipswich, MA, USA).
3.5. Synthesis of RNA Transcripts
Calculation of copy numbers and synthesis of RNA transcripts were performed as described in previous studies (11, 12). Three types of RNA transcripts were prepared for assays: real-time RT-LAMP, conventional RT-PCR, and real-time RT-PCR; each transcript contained the primer sequence regions. RNA transcripts were used as templates after 10-fold serial dilutions from 5 × 106 down to 5 × 101 copies. The efficiency and sensitivity of the three assays were evaluated.
4. Results
4.1. Specificity of The Real-Time RT-LAMP Assay
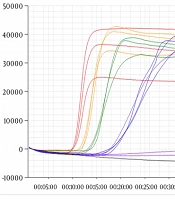
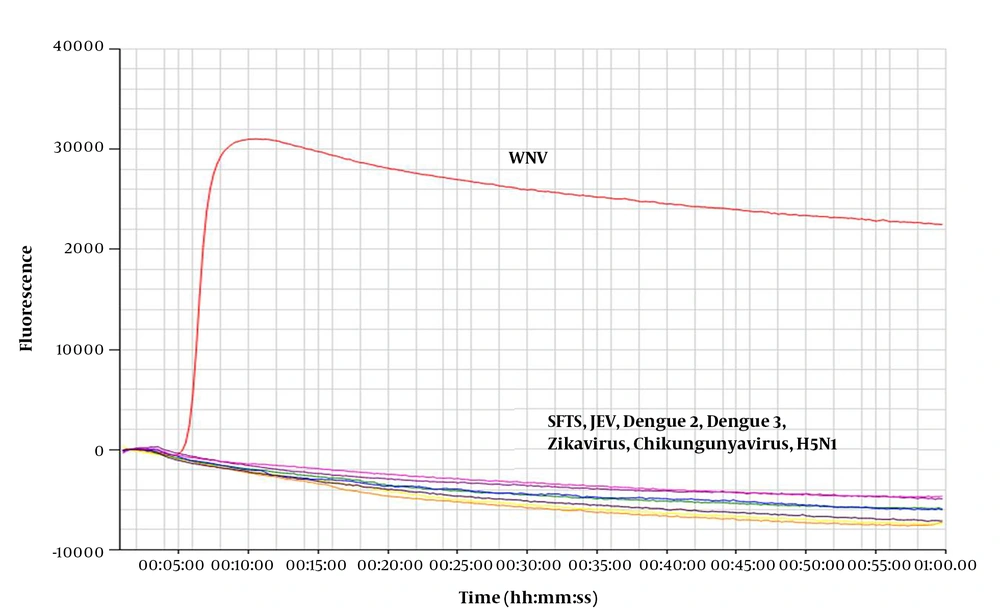
For the specificity test, extracted viral RNA was used as template for the real-time RT-LAMP assay. Novel primers were used for detection (Table 1). The results were positive for WNV, while SFTSV, JEV, DENV-2, DENV-3, Zika virus, chikungunya virus, and H5N1 were not amplified (Figure 1). These results indicate that the novel primer for the real-time RT-LAMP assay can detect WNV with high specificity.
RNA extracted from West Nile Virus (WNV) was amplified by the real-time RT-LAMP assay, however no products were amplified from the control viral RNA. Control viruses: Zika virus, chikungunya virus, influenza virus (H5N1), severe fever with thrombocytopenia syndrome virus (SFTSV), Japanese encephalitis virus (JEV), and dengue viruses 2 and 3 (DENV-2, DENV-3).
4.2. Sensitivity and Efficiency Test for WNV Detection
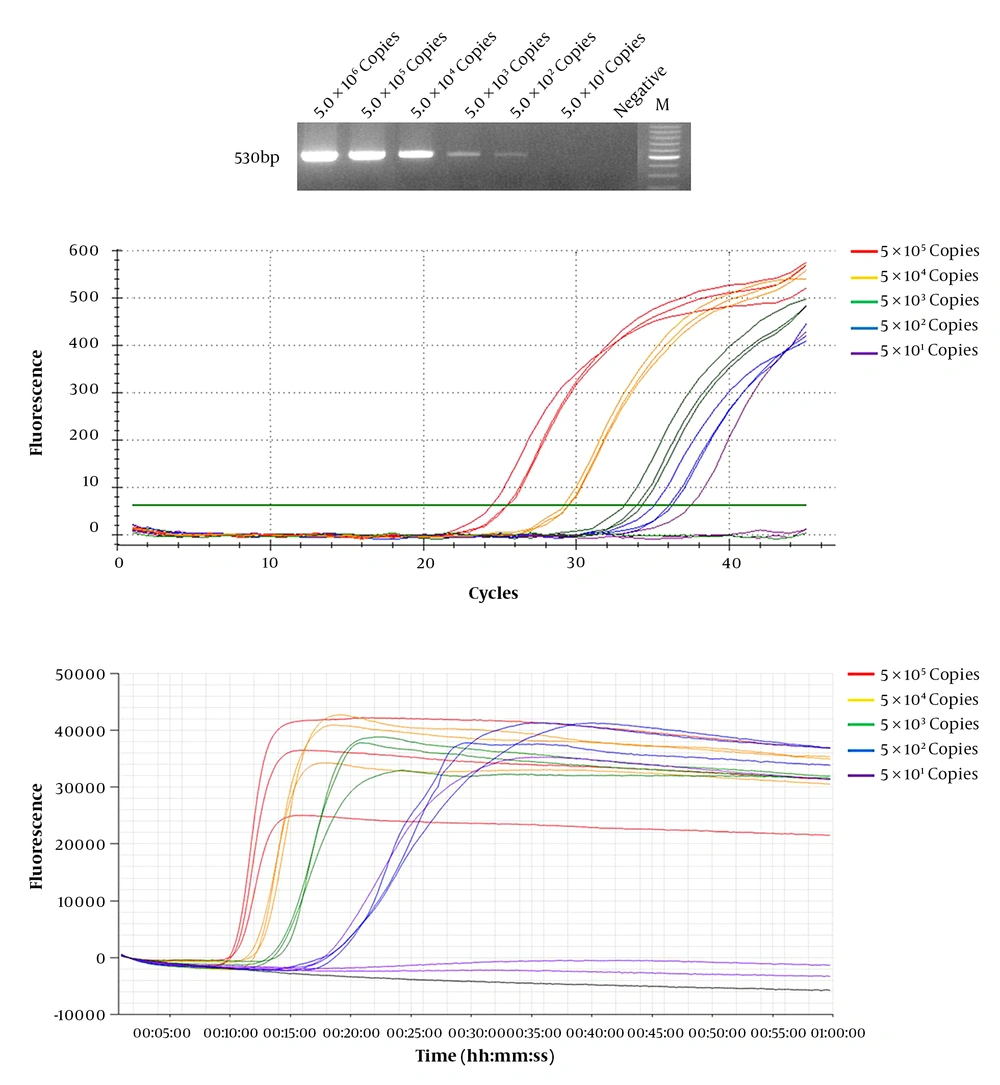
We compared the efficiency and sensitivity of WNV detection in conventional RT-PCR, real-time RT-PCR, and real-time LAMP assay. Known concentrations of diluted WNV RNA transcripts (5 × 101 – 5 × 106 copies) were used as templates. Real-time RT-PCR and real-time LAMP assays were used to amplify the targeted RNA with a detection limit of 5 × 101 copies (Table 2). The detection limit of conventional RT-PCR was 5 × 102 copies (Figure 2). These results show that real-time RT-LAMP is more sensitive than conventional RT-PCR. Real-time RT-LAMP assay is more efficient than conventional RT-PCR and real-time RT-PCR, and provided results within 9 to 18 min. In contrast, real-time RT-PCR provided results after 35 min (21 to 35 cycles) at the same WNV RNA copy number (Table 2). Moreover, conventional RT-PCR required an additional 6 h to yield results after performing a second PCR using agarose gel.
Diluted West Nile Virus (WNV) RNA was used for detection sensitivity and efficiency tests. Serial dilution of WNV RNA, ranging between 5 × 106 and 5 × 101 were evaluated by (A) conventional RT-PCR, (B) real-time RT-PCR, and (C) real-time RT-LAMP assay, and detections limit was 5 × 102, 5 × 101, and 5 × 101 copies respectively. The real-time LAMP assay showed results in 9 – 18 min; however, the other two methods took at least 35 min to over 6 h.
| Template Concentration | Real-Time RT-PCR, Time a(Cycle) | Real-Time LAMP Assay, Time |
|---|---|---|
| 105 | 25:28/26:00/26:14, 21.69/22.51/22.86 | 9:54/9:54/10:05 |
| 104 | 28:20/28:41/28:49, 26.01/26.53/26.73 | 11:16/11:35/11:58 |
| 103 | 30:32/31:33/32:02, 29.31/30.83/31.55 | 13:05/13:29/13:42 |
| 102 | 31:39/32:32/33:18, 30.97/32.31/33.44 | 17:31/17:58/18:36 |
| 101 | 34:19/N. D/N. D, 34.98/N. D/N. D | 17:42/N. D/N. D |
Abbreviation: N.D, no data.
a Time (mm:ss).
5. Discussion
Since its emergence in New York City in 1999, WNV has become endemic in the United States (6, 13). WNV has annual seasonal outbreaks. Most commonly, patients are asymptomatic but fever symptoms occurs in 20% of the population, and < 1% progress to WNND, which might include acute flaccid paralysis, meningitis, and encephalitis (6). The research group estimates average costs for treating hospitalized WNV patients to be US $56 million annually and US $700,000 per patient for initial and long-term cases (14, 15). WNV burdens many countries, not only the United States (2, 16-18). To reduce the WNV burden and outbreaks, a quick and accurate diagnostic method is required. The conventional diagnosis methods used ELISA and quantitative real-time RT-PCR (19-24). These methods target the E gene or a neutralizing epitope within the envelope such as E glycoprotein. In this study, we designed a primer targeting the most conserved region of the E gene. ELISA, conventional RT-PCR, and real-time RT-PCR have limitations such as being time-consuming, low sensitivity, and being highly technical. LAMP assay is a simple, rapid, and accurate method that can be used for the detection of several RNA viruses including WNV (25-29). Our results show that the LAMP assay is about 10-fold more sensitive than conventional RT-PCR.
The results also show that it is much more efficient and rapid than conventional RT-PCR and real-time RT-PCR, which take at least 35 min to 6 h and require an additional step for confirmation. However, by using the novel designed primers for LAMP assay we were able to confirm the presence of WNV in 18 min. We interpret the detection of Real-time RT-LAMP and real-time RT-PCR as 5 × 101 copies, because the limit reaction is the minimum detection limit. WNV poses a threat to public health in many countries around the world and is also an economic burden. This virus has undergone a rapid geographical spread over the past few decades (14, 30). To reduce this, rapid and efficient diagnostic methods must be developed. This study found the LAMP assay to be the best diagnostic method for WNV detection. We expect our methods to be helpful to researchers and clinicians.
5.1. Conclusions
In conclusion, novel primers were developed by analyzing of 32 genomic sequences of WNV strains. The primers were designed from the most conserved region of the E gene for real-time RT-LAMP. The LAMP assay is a rapid, efficient, highly sensitive, and specific tool for the identification of WNV. It will be useful for quick detection of WNV infection and will contribute to the epidemiology study.