1. Background
Nontuberculous mycobacteria (NTM) are a group of opportunistic pathogens, which are ubiquitously found in the environment. Considering the median growth rate, NTM species are classified into slowly growing mycobacteria (SGM) and rapidly growing mycobacteria (RGM). In recent decades, with the emergence of immunodeficiency diseases, the incidence of NTM infection has been rising (1). Besides, the isolation rate of NTM has been gradually increasing, as shown in an epidemiological tuberculous (TB) study from China (4.3% in 1979, 11.1% in 2000, and 21% in 2010) (2).
The NTM infection has similar clinical, radiological, and pathological features to TB. The common sputum culture test by acid-fast bacilli (AFB) cannot effectively distinguish NTM infection from TB, and many NTM patients are misdiagnosed with TB (3, 4). In terms of treatment, NTM infection, due to its cell wall permeability barrier and highly hydrophobic surface, shows natural resistance to most anti-TB drugs. Also, different types of NTM, with distinct characteristics, show different susceptibilities to drugs (5). China is one of the top ten countries in terms of the TB burden worldwide. Besides, NTM infection increases the challenges of diagnosis and treatment of TB (6). However, culture and identification of NTM infection are not routinely performed in most regions of China, and the exact burden of NTM infection remains unknown (7).
2. Objectives
This study was done to retrospectively analyze the prevalence, identification, DST results, and clinical data of NTM infection among TB suspects in Northwest China to facilitate NTM management.
3. Methods
3.1. Study Subjects
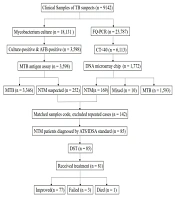
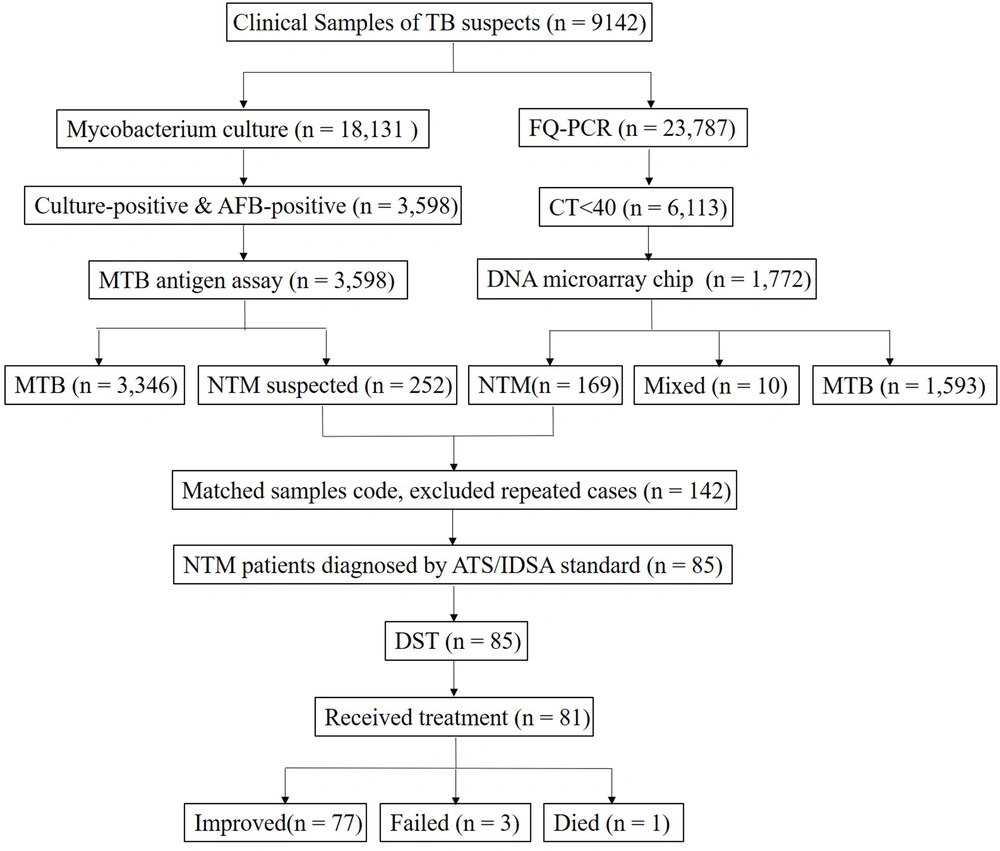
Xi’an Chest Hospital treats TB suspects from local areas and four surrounding provinces. Among TB suspects admitted to this hospital from January to December 2020, 169 cases with NTM diagnosis data were examined and also, the DST and clinical data of 85 NTM patients were collected and analyzed. Sample collection and diagnosis were based on the American Thoracic Society (ATS) guidelines (8). The processes of data collection and matching are presented in Figure 1. The demographic, clinical, and radiological data were collected from the electronic medical records system.
The flow diagram of data collection and procedures. The samples were divided into two parts for molecular biology techniques and culture, respectively. After matching the codes of identification and culture results, nontuberculous mycobacteria (NTM) patients were diagnosed based on the standard. Then, drug-sensitive test (DST) was performed for the NTM patients.
3.2. Mycobacterial Culture and Characterization
Samples of early morning sputum, bronchoalveolar fluid, pleuroperitoneal fluid, purulence, tissue, and urine were collected in this study. According to the ATS guidelines, two samples obtained on different occasions were tested separately; potentially contaminated samples were excluded. The samples were divided into two parts; one part was tested using molecular biology techniques (PCR-fluorescent probe and DNA microarray), and the other part was cultured.
3.2.1. Nontuberculous Mycobacteria Identification by PCR-Fluorescent Probe and DNA Microarray
The samples were liquefied with 4% sodium hydroxide, centrifuged, and rinsed twice with 0.9% sodium chloride. Next, they were mixed with 50 µL of nucleic acid extraction reagent and swirled. Nucleic acid was extracted using an ExtractorTM 34 system (Bo’ao Biotechnology Co., Ltd, Beijing, China). Then, 2 µL of the nucleic acid solution was amplified and detected using an ABI 7500 PCR system (BD Diagnostic Systems, Sparks, Maryland, USA) (9). If the cycle threshold (Ct) in PCR was less than 40, nucleic acid was identified by DNA microarray chip (Bo’ao Biotechnology Co., Ltd., Beijing, China) (10).
3.2.2. Nontuberculous Mycobacteria Culture and Drug Susceptibility Testing
The samples were simultaneously treated with both 1% N-acetyl-L-cysteine (NALC) and 4% sodium hydroxide. The remaining sediment was suspended in sterile phosphate-buffered saline (pH 6.8) and vortexed. The mixture was dissolved and blended with a nutritional additive growth supplement in a PANTA reagent bottle. The processed samples were added to the MGIT liquid culture tube and incubated using a BACTEC MGIT 960 system (BD Diagnostic Systems, Sparks, Maryland, USA) at 37°C. Some SGM that could not grow at 35°C to 37°C, were re-cultured at 30°C (11). The positive samples were tested by AFB smear microscopy and rapid TB antigen assay. The MPB64 monoclonal antibody (Innovative Biological Technology Co., Ltd., Hangzhou, China) was used as the TB antigen. The microscopy-positive, but TB antigen-negative samples, were considered as NTM infection suspects.
After matching the codes of identified NTM samples and cultured NTM-suspected samples, the identification results were used to diagnose NTM infection. The isolated strains of NTM patients were tested for susceptibility to 15 drugs with a YK909 MicroDSTTM instrument (Yingke Biotechnology Co., Ltd., Zhuhai, China). The tested drugs included: rifampicin (Rfp), ethambutol (Emb), moxifloxacin (Mfx), amikacin (Am), clarithromycin (Clr), rifabutin (Rfb), linezolid (Lzd), imipenem and cilastatin (Lmp/Cln), azithromycin (Azm), cefoxitin (Fox), tobramycin (Tob), gatifloxacin (Gfx), doxycycline (Dox), minocycline (Min), and sulfamethoxazole (Smz). Further details about the concentration gradients of the drugs are presented as supplementary information (Appendix 1).
3.3. Statistical Analysis
Normally distributed continuous variables are presented as mean ± SD. The groups were compared using the t-test. Nonnormally distributed variables are presented as median (Q1 - Q3) and examined by the Mann-Whitney U test. Besides, categorical variables in 2 × 2 tables were analyzed using the Chi-square test or continuity correction test (1 < T < 5). The Chi-square test was also performed for likelihood ratio testing of variables in R × C tables. The level of statistical significance was considered to be P < 0.05, and data were interpreted at 95% confidence intervals (CIs). SPSS version 22.0 (SPSS Inc., Chicago, USA) was used for data analysis.
4. Results
4.1. Nontuberculous Mycobacteria Species Distribution
From January to December 2020, a total of 23,787 clinical specimens were evaluated for mycobacteria tuberculous (MTB)/NTM infections using PCR-fluorescent probe assays, and overall, 6,113 samples (25.7%) were positive (Ct < 40). Among positive samples, 1,772 non-repeat nucleic acid samples were identified by DNA microarrays, including 1593 (89.9%) TB complex samples, 169 (9.5%) NTM samples, and 10 (0.6%) TB + NTM samples. The species and sample sources of 169 NTM strains are presented in Table 1. The most common species were Mycobacterium chelonae/ M. abscessus (64/169, 37.7%), and M. intracellulare (40/169, 23.7%). The most common samples were sputum (117/169, 69.2%) and lavage fluid (40/169, 23.7%). Also, M. chelonae/M. abscessus showed the highest frequency (48/117, 41.0%) in sputum. In bronchoalveolar fluid samples, most strains were identified as M. intracellulare (15/40, 37.5%).
| Sample Sources | Rapidly Growing Mycobacteria | Slowly Growing Mycobacteria | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M. chelonae/ M. abscessus | M. fortuitum | M. phlei | M. smegmatis | M. intracellulare | M. kansassi | M. avium | M. gordonae | M. gilvum | M. terrae | Total | |
| No. | 64 | 10 | 1 | 2 | 40 | 25 | 16 | 8 | 2 | 1 | 169 |
| Sputum | 48 (75.0) | 7 (70.0) | 1 (100.0) | 1 (50.0) | 24 (60.0) | 12 (48.0) | 15 (93.7) | 6 (75.0) | 2 (100.0) | 1 (100.0) | 117 (69.2) |
| Bronchoalveolar fluid | 9 (14.1) | 1 (10.0) | 0 | 1 (50.0) | 15 (37.5) | 12 (48.0) | 1 (6.3) | 1 (12.5) | 0 | 0 | 40 (23.7) |
| Pleuroperitoneal fluids | 1 (1.6) | 0 | 0 | 0 | 1 (2.5) | 0 | 0 | 1 (12.5) | 0 | 0 | 3 (1.8) |
| Purulence | 2 (3.1) | 2 (20.0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 (2.4) |
| Tissue | 3 (4.7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (1.8) |
| Urine | 1 (1.6) | 0 | 0 | 0 | 0 | 1 (4.0) | 0 | 0 | 0 | 0 | 2 (1.2) |
4.2. Drug susceptibility Testing of Nontuberculous Mycobacteria Strains
From January to December 2020, 18,131 clinical samples were cultured using a BACTEC MGIT 960 system. After testing via AFB microscopy and rapid TB antigen assay, 252 strains were considered as NTM suspects. By matching the sample codes and excluding repeated cases, 142 samples were cultured and identified simultaneously. The susceptibility of 85 isolated strains to 15 drugs was tested by DST. As illustrated in Table 2, all strains were resistant to Lmp/Cln (85/85, 100%), while most strains were sensitive to Lzd (4.7%, 4/85). The RGM strains were resistant to 7.7 drugs on average, while the SGM strains were resistant to 3.9 drugs on average (t = 6.66, P < 0.001). Only five strains showed resistance to more than 11 drugs. For the majority of identified strains, there were several medication regimens. As shown in Table 2, different strains, especially RGM and SGM, showed discrepant susceptibility.
| Drugs | Rapidly Growing Mycobacteria | Slowly Growing Mycobacteria | ||||||
|---|---|---|---|---|---|---|---|---|
| M. chelonae/ M. abscessus | M. fortuitum | M. intracellulare | M. kansassi | M. avium | M. gordonae | M. gilvum | Total | |
| No. | 30 | 5 | 26 | 13 | 7 | 2 | 2 | 85 |
| Any drug resistance | ||||||||
| Rfp | 22 (73.3) | 1 (20.0) | 2 (7.7) | 2 (15.4) | 0 | 0 | 0 | 27 (31.8) |
| Emb | 28 (93.3) | 3 (60.0) | 3 (11.5) | 1 (7.7) | 0 | 0 | 1 (50.0) | 36 (42.4) |
| Mfx | 10 (33.3) | 0 | 0 | 0 | 0 | 0 | 1 (50.0) | 11 (12.9) |
| Am | 1 (3.3) | 0 | 0 | 3 (23.1) | 0 | 0 | 0 | 4 (4.7) |
| Clr | 4 (13.3) | 2 (40.2) | 0 | 1 (7.7) | 0 | 0 | 0 | 7 (8.2) |
| Rfb | 11 (36.7) | 0 | 1 (3.9) | 1 (7.7) | 0 | 0 | 0 | 13 (15.3) |
| Lzd | 1 (3.3) | 1 (20.0) | 0 | 1 (7.7) | 1 (14.3) | 0 | 0 | 4 (4.7) |
| Lmp/Cln | 30 (100.0) | 5 (100.0) | 26 (100.0) | 13 (100.0) | 7 (100.0) | 2 (100.0) | 2 (100.0) | 85 (100.0) |
| Azm | 13 (43.3) | 4 (80.0) | 0 | 4 (30.8) | 1 (14.3) | 0 | 1 (50.0) | 23 (27.1) |
| Fox | 1 (3.3) | 0 | 1 (3.9) | 10 (76.9) | 0 | 0 | 0 | 12 (14.1) |
| Tob | 22 (73.3) | 5 (100.0) | 2 (7.7) | 12 (92.3) | 0 | 1 (50.0) | 2 (100.0) | 46 (54.1) |
| Gfx | 28 (93.3) | 0 | 10 (62.5) | 9 (69.2) | 0 | 1 (50.0) | 1 (50.0) | 49 (57.6) |
| Dox | 23 (76.7) | 4 (80.0) | 15 (57.7) | 10 (76.9) | 1 (14.3) | 0 | 1 (50.0) | 54 (63.4) |
| Min | 20 (66.7) | 4 (80.0) | 14 (53.9) | 5 (38.5) | 3 (42.9) | 0 | 1 (50.0) | 47 (55.3) |
| Smz | 26 (86.7) | 3 (60.0) | 13 (50.0) | 12 (92.3) | 2 (28.6) | 1 (50.0) | 2 (100.0) | 59 (69.4) |
| Monoresistance | ||||||||
| Lmp/Cln | 0 | 0 | 6 (23.1) | 0 | 3 (42.9) | 1 (50.0) | 0 | 10 (11.8) |
| 2 ~ 4 drugs | 5 (16.7) | 1 (20.0) | 16 (61.5) | 4 (30.8) | 4 (57.1) | 1 (50.0) | 1 (50.0) | 32 (37.6) |
| 5 ~ 7 drugs | 6 (20.0) | 2 (40.0) | 3 (11.5) | 5 (38.5) | 0 | 0 | 0 | 16 (18.8) |
| 8 ~ 10 drugs | 15 (50.0) | 2 (40.0) | 1 (3.8) | 3 (23.1) | 0 | 0 | 1 (50.0) | 22 (25.9) |
| ≥ 11 drugs | 4 (13.3) | 0 | 0 | 1 (7.7) | 0 | 0 | 0 | 5 (5.9) |
Abbreviations: Rfp, rifampicin; Emb, ethambutol; Mfx, moxifloxacin; Am, amikacin; Clr, clarithromycin; Rfb, rifabutin; Lzd, linezolid; Lmp/Cln, imipenem and cilastatin; Azm, azithromycin; Fox, cefoxitin; Tob, tobramycin; Gfx, gatifloxacin; Dox, doxycycline; Min, minocycline; Smz, sulfamethoxazole.
aValues are expressed as No. (%) unless otherwise indicated.
4.3. Demographic and Laboratory Characteristics of Nontuberculous Mycobacteria Patients
According to the ATS definition, 85 patients were diagnosed with an active NTM infection, based on the clinical, radiological, and pathogenic features. The mean age of the subjects was 49.4 years (range: 17 - 85 years). The age distribution of the patients was as follows: 30 cases in the age range of 21 - 40 years; 25 cases in the age range of 41 - 60 years; and 28 cases in the age range of > 61 years. There was no significant difference in the mean age of male and female subjects (t = 0.021, P = 0.841). Compared to the RGM strains, the SGM strains were mostly identified in male subjects (P = 0.009); however, they were less frequent in T-SPOT-positive patients (P = 0.004) and were associated with fewer inpatient days (P = 0.020). In the TB protein chip test, five antibodies (16 kDa, 38 kDa, LAM, Rv1636, and CFP10) were evaluated, one of which showed a positive result. The two antibodies with the highest positive rates were 38 kDa (19/85, 22.4%) and LAM (17/85, 20%) (Table 3).
| Total | RGM | SGM | t | P | |
|---|---|---|---|---|---|
| Male, No. (%) | 46 (54.1) | 13 (37.1) | 33 (66.0) | 6.91 | 0.009 b |
| Age, y, median (Q1 ~ Q3) | 53 (31-65) | 48 (32-65) | 55 (30-67) | - | 0.544 c |
| Inpatient days | 50.8 ± 30.2 | 60.7 ± 36.7 | 43.9 ± 22.7 | 2.40 | 0.020 |
| Occupation, No. (%) | |||||
| Famer & worker | 49 (57.6) | 22 (62.9) | 27 (54.0) | 2.920 | 0.232 d |
| Student & office clerk | 19 (22.4) | 9 (25.7) | 10 (20.0) | ||
| Unemployed & retiree | 17 (20.0) | 4 (11.4) | 13 (26.0) | ||
| T-SPOT POS, No. (%) | 33 (38.8) | 20 (57.1) | 13 (26.0) | 8.407 | 0.004 b |
| TB protein chip POS, No. (%) | 37 (43.5) | 19 (54.3) | 18 (36.0) | 2.801 | 0.094 b |
| Diabetes, No. (%) | 7 (8.2) | 2 (5.7) | 5 (10.0) | 0.094 | 0.759 |
| Diagnosis, No. (%) | |||||
| Pulmonary | 78 (91.8) | 30 (85.7) | 48 (96.0) | 1.68 | 0.195 |
| Extrapulmonary | 7 (8.2) | 5 (14.3) | 2 (4.0) | ||
| Radiological features of pulmonary NTM | 78 | 30 | 48 | ||
| Bronchiectasis | 74 (94.9) | 28 (93.3) | 46 (95.8) | 0.237 | 0.626 b |
| Cavity | 26 (33.3) | 11 (36.7) | 15 (31.3) | 0.244 | 0.621 b |
| Nodules | 15 (19.2) | 9 (30) | 6 (12.5) | 3.640 | 0.056 b |
| Consolidation | 6 (7.7) | 2 (6.7) | 4 (8.3) | 0.072 | 0.788 b |
| Collapse/atelectasis | 4 (8.9) | 2 (6.7) | 2, 4.2% | 0.237 | 0.626 b |
| Ground glass opacity | 1 (1.3) | 1 (3.3) | 0 | 1.621 | 0.203 b |
Abbreviations: NTM, nontuberculous mycobacteria; TB, tuberculous mycobacteria; POS, positive; RGM, rapidly growing mycobacteria; SGM, slowly growing mycobacteria.
aValues are expressed as No. (%) unless otherwise indicated.
b Chi-square test.
c Mann–Whitney U test.
d likelihood ratio Chi-square tested continuity correction test.
4.4. Clinical Characteristics of Nontuberculous Mycobacteria Patients
The mean length of inpatient hospital stay was 50.9 days (range: 14 - 154 days) in 85 NTM patients. Of these 85 patients, 78 cases (78/85, 91.5%) were diagnosed with NTM lung disease, while 7 cases (7/85, 8.2%) were diagnosed with an extrapulmonary NTM infection (three lymph node samples, two skin samples, one urinary sample, and one mammary gland sample). Among 78 patients with NTM lung disease, 19 cases (24.4%) were misdiagnosed with TB, and 17 cases (21.8%) were misdiagnosed with pneumonia or bronchitis. Their clinical respiratory symptoms were as follows: coughing with sputum in 68 cases (87.2%), shortness of breath in 31 (36.5%) cases, fever in 27 (31.8%) cases, hemoptysis in 13 (15.3%) cases, fatigue in 8 (9.4%) cases, and anorexia in 28 (34.1%) cases. Also, 26 patients (33.3%) had a TB history, while three patients (3.85%) had an NTM infection history. Except for four patients who did not complete intensive therapy during their hospitalization due to financial reasons, 81 patients were treated with drug regimens according to the identified NTM species and DST results. After hospitalization, 77 patients (95.1%) improved clinically, 3 patients (3.7%) showed treatment failure, and 1 patient (1.2%) died (Figure 1).
4.5. Radiological Features of Nontuberculous Mycobacteria Lung Disease
All 78 patients with NTM lung disease were observed in terms of radiological changes according to the nodular distribution patterns on chest computed tomography (CT) scans. The lesion sites (12) included the lung field I (in both lungs, posterior apical and inferior dorsal lobes) in 53 patients (67.9%), lung field II (in the middle, tongue segment) in 41 patients (52.6%), lung field III (the anterior segment of upper lobe or basal segment of the lower lobe) in 26 patients (33.3%), and diffuse distribution (diffuse in both lung fields) in 2 patients (2.6%). The manifestations are shown in Table 3.
5. Discussion
The NTM are widely distributed in the environment. Most of these bacteria were previously considered to be non-pathogenic (13). According to multiple studies, the NTM infection is increasing globally, and some cases are very difficult to treat (14, 15). It is known that different NTM species can be found in extremely different geographical areas. For example, almost 80% of NTM lung disease cases in the US were reportedly caused by Mycobacterium avium (16); the corresponding rates were 43% in the UK (17), 65% in South Korea (18), and 56% in Asia (19).
The prevalence of NTM infection varies greatly across China. Multicenter studies have indicated that regions with higher humidity exhibit a higher prevalence of NTM infection (2, 20, 21). The NTM lung disease and multidrug-resistant TB can be easily misdiagnosed due to similar clinical symptoms, pathological changes, radiological features, and several antibody test results (22, 23). Because of the dispersant antibiotic susceptibility of NTM species, besides a pervasive and steady increase in NTM infections, timely and accurate identification of mycobacterial species has become one of the main challenges of clinical laboratories (24).
The present study identified NTM species rapidly by performing molecular diagnostics and explored the feasibility of several antibiotics for NTM treatment. In comparison with the traditional proportional DST, the MicroDSTTM assay could provide more data in a shorter period. According to the DST results and previous studies, LMP/Cln (100% drug resistance) should be replaced by clofazimine, protionamide, and other new drugs, such as bedaquiline (13, 25). Also, to accelerate antidiastole, multi-slice spiral CT (MSCT) scan can be used as a routine examination for NTM suspects (26).
Unlike previous reports from northeast and south of China (27), which showed that the number of NTM patients increased with age, in this study, the most affected age group was 21 - 40 years. This finding may be related to the selection of research subjects (TB suspects) because patients identified as TB positive in the same period were in a similar age range. Based on the present study, it can be concluded that the characteristics of RGM strains are more important than SGM strains, as these strains showed resistance to more drugs and were associated with longer inpatient admission. Also, in terms of risk factors, females were more likely to be infected with RGM, and patients with RGM strains tended to have positive T-SPOT test results. However, to investigate the influential factors in more depth, further research is needed by increasing the sample size.
5.1. Conclusions
Among TB suspects in Northwest China, the NTM species with the highest isolation rates were M. chelonae/M. abscessus and M. intracellulare. Different strains, especially SGM and RGM, showed different characteristics and discrepant susceptibility to different drugs. Because the rapidly identified strains on molecular detection tests can be administered to NTM patients to accelerate treatment, after this therapy, the treatment regimen can be adjusted according to the minimum inhibitory concentration (MIC) of the MicroDSTTM test. Overall, timely identification and accurate DST of NTM species are crucial for managing NTM infections.