1. Background
Antimicrobial resistance (AMR) has become a significant problem with the increasing burden for healthcare systems worldwide. In recent years, antibiotic resistance has been associated with considerable morbidity and mortality rates due to prolonged hospitalization (1). Although the occurrence of antibiotic resistance is a natural phenomenon in bacteria, it is exacerbated by the misuse of antibiotics in humans and animals (2). A significant reason for the rapid spread of antibiotic resistance is the highly mobile genetic elements. These elements can replicate and pass among bacterial species (3). Acinetobacter baumannii is a common nosocomial pathogen that causes different infections, such as ventilator-associated pneumonia, bacteremia, urinary tract infections, surgical site infections, and secondary meningitis in hospitalized patients, especially those with immunodeficiency (4). The relatively recent emergence and increased prevalence of multidrug-resistant (MDR) A. baumannii has been an issue of great concern. The World Health Organization (WHO) lists A. baumannii as a critical pathogen, highlighting the need to develop new and effective antibiotics (5).
Miserably, the number of MDR A. baumannii isolates has increased significantly worldwide. Also, A. baumannii can acquire or upregulate resistance genes through genomic plasticity, limiting effective therapeutic options and increasing mortality rates. This phenomenon can increase resistance to multiple antibiotics, including those used as a last resort, such as carbapenems, reserved for cases where all alternatives have been depleted (6). The enzymes IMP, VIM, GIM, SIM, and NDM are classified as class (B) Metallo-β-lactamases (MBLs), and their genes are mostly found in transmissible plasmids (7).
In Acinetobacter spp., the acquisition and spread of an antimicrobial-resistant determinant in hospitals and communities are often facilitated by horizontal gene transfer of mobile genetic elements, including plasmids, transposons, and integrons. Among these mobile elements, integrons are unique for their ability to carry and express resistance genes (8). It has been suggested that multidrug-resistant strains acquire their antibiotic-resistant genes via integrons that take single or multiple gene cassettes (9). Integrons carrying different cassette arrays have been reported in several studies from South America to Far East Asia (10).
2. Objectives
The current study aimed at characterizing class 1, 2, and 3 integrons among clinical carbapenemase-producing A. baumannii isolates from hospitalized patients in Tehran, Iran, followed by the genotypic analysis of these isolates.
3. Methods
3.1. Collection and Identification of Bacterial Isolates
In this cross-sectional study, a total of 103 consecutive non-duplicate A. baumannii isolates collected from clinical specimens in hospitalized patients from an educational hospital in Tehran, Iran, from November 2019 to July 2020 were investigated. The isolates were obtained from sputum, tracheal aspirate, wound, catheter, and cerebrospinal fluid (CSF). Standard microbiological and biochemical tests, including triple sugar iron agar (TSI), indole, methyl red (MR), Voges–Proskauer (VP), citrate (IMVIC), and oxidase test were used to identify A. baumannii isolates. All K/K colonies on TSI and oxidase negative coccobacilli (11) were genotypically confirmed as A. baumannii by the presence of the blaOXA-51-like (1) and rpoB PCR sequencing (12).
3.2. Antibiotic Susceptibility Testing
According to the Clinical and Laboratory Standards Institute (CLSI), the antibiotic susceptibility profiles of A. baumannii isolates were determined using the disk diffusion method. In this step, the results were interpreted with criteria published in CLSI 2019 (13). For this purpose, various antibiotic disks, such as ampicillin/sulbactam (SAM, 10 µg), minocycline (MN, 30 µg), meropenem (MEM, 10 µg), amikacin (AN, 30 µg), ciprofloxacin (CIP, 5 µg), trimethoprim-sulfamethoxazole (SXT, 1.25 + 23.75 µg), and ceftazidime (CAZ, 30 µg) were used. In the next step, minimum inhibitory concentrations (MICs) were determined for imipenem in carbapenem-resistant isolates using E-test (bioMérieux). Antibiotic susceptibility was interpreted based on CLSI clinical breakpoints. The Escherichia coli ATCC 25922 was used as a quality control strain. The categorizations of multidrug-resistant (MDR), extensively drug-resistant (XDR), and pan-drug resistant (PDR) A. baumannii were performed based on Magiorakos criteria (14).
3.3. Detection of Genes Encoding β-lactamase, Integrases, and Clonal Complex Analysis
DNAs of bacterial isolates were extracted as previously described (15). A PCR experiment was used to determine the presence of genes producing carbapenemases and integrases using primers targeting blaOXA-23-like, blaVIM, blaNDM, intI1, intI2, and intI3 genes (Table 1). The PCR conditions were based on the mentioned reference (16). All isolates were confirmed as A. baumannii by sequencing of the blaOXA-51-like, an intrinsic enzyme marker, and ropB, as described previously (15). Determination of the allele number and detection of the clonal complex (CC) for each isolate were performed by a combination of sequence-based typing (SBT) of blaOXA-51-like and ampC as reported previously (15).
| Target | Primer Sequence (5ˊ - 3ˊ) | Product Size (bp) | Reference |
|---|---|---|---|
| rpoB | 1024 | (12) | |
| F | CTGACTTGACGCGTGA | ||
| R | TGTTTGAACCCATGAGC | ||
| blaOXA-51-like | 501 | (17) | |
| F | GATCGGATTGGAGAACCAGA | ||
| R | ATTTCTGACCGCATTTCCAT | ||
| blaNDM | 155 | (18) | |
| F | GCGCAACACAGCCTGACTTT | ||
| R | CAGCCACCAAAAGCGATGTC | ||
| blaVIM | 518 | (19) | |
| F | GGGAGCCGAGTGGTGAGT | ||
| R | GGCACAACCACCGTATAG | ||
| intI1 | 250 | (20) | |
| F | TCTCGGGTAACATCAAGG | ||
| R | AGGAGATCCGAAGACCTC | ||
| intI2 | 789 | (21) | |
| F | CACGGATATGCGACAAAAAGGT | ||
| R | GTAGCAAACGAGTGACGAAATG | ||
| intI3 | 980 | (21) | |
| F | GCCTCCGGCAGCGACTTTCAG | ||
| R | ACGGATCTGCCAAACCTGACT | ||
| Conserved segment of class 1 integrons | Variable | (21) | |
| 5′-CS | GGCATCCAAGCAGCAAG | ||
| 3′-CS | AAAGCAGACTTGACCTGA or GAAGCGGCGTCGGCTTGA | ||
| Conserved segment of class 2 integrons | Variable | (21) | |
| 5′-CS | ACCTTTTTGTCGCATATCCGTG | ||
| 3′-CS | TACCTGTTCTGCCCGTATCT |
3.4. PCR Amplification and Sequencing of Integrons Internal Variable Region
All integron–positive MDR A. baumannii strains were assessed for variable regions of Class 1 - 2 integrons by the primers 5′-CS/3′-CS. The PCR conditions were based on the mentioned reference (9). Sequencing of the purified PCR amplicons was performed using DNA analyzers (Applied Biosystems, Inc.). The nucleotide sequence analysis was performed by the BLAST tool at the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (22). The sequences were manually analyzed using CLC main workbench software version 20 (CLC Bio, Aarhus, Denmark).
3.5. Statistical Analyses
The normality of continuous data distribution was assessed by the Kolmogorov-Smirnov test. Numerical data were summarized as means and standard deviations or median and interquartile range as appropriate. Categorical data were summarized as frequencies and proportions. The association of variables was analyzed using one-way analysis of variance, student t-test, and Mann-Whitney U Kruskal-Wallis tests as appropriate. All statistical analyses were conducted with STATA 12.0 (StataCorp LP, College Station, TX, USA), SPSS for Windows version 24.0 (IBM Corp., Armonk, NY), and GraphPad Prism software version 8.0 (GraphPad Software Inc., La Jolla, CA, USA). A P < 0.05 was defined as statistical significance in all tests.
4. Results
4.1. Collection and Identification of Bacterial Isolates
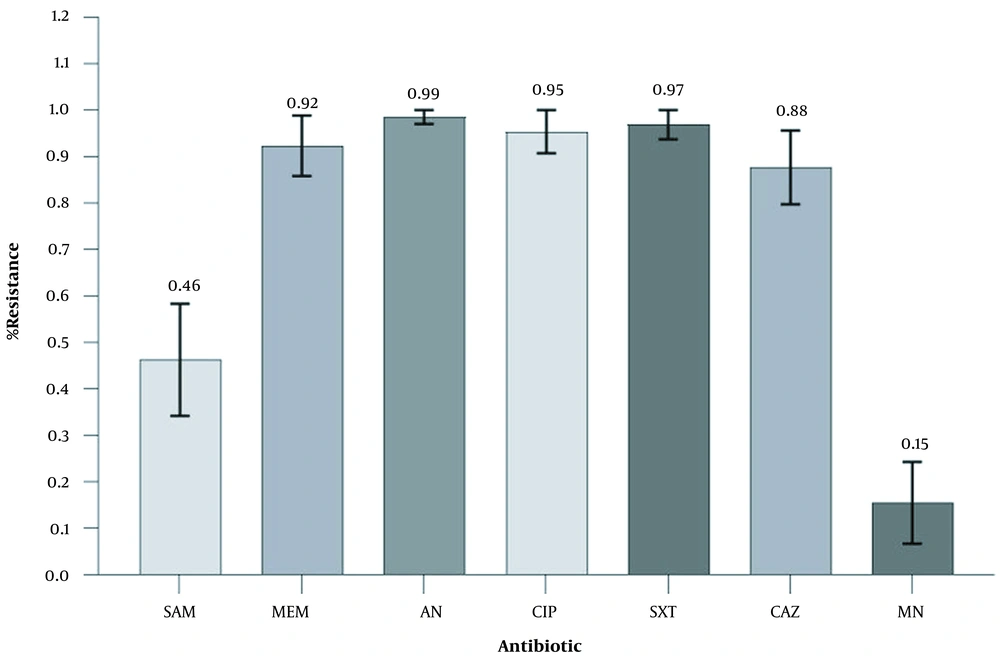
A total of 65 non-repetitive isolates were collected and confirmed as MDR A. baumannii by phenotypic and genotypic antimicrobial methods. The isolates were recovered from clinical specimens, including tracheal aspirate (n = 57/65; 87.69%), sputum (n = 3/65; 4.62%), catheter (n = 3/65; 4.62%), CSF (n = 1/65; 1.54%), and wound (n = 1/65; 1.54%). The mean age of the patients was 48.25 ± 21.09 years (ranging from 5 to 96). Also, 41 patients (63.1%) were male, and 24 cases (36.9%) were female (Table 2). Antimicrobial susceptibility results for the 65 A. baumannii isolates are shown in Table 2. The isolates showed MDR phenotypes and resistance to most of the tested antibiotics by the disk diffusion method, in particular high-level resistance to AN (n = 64; 98.46%), SXT (n = 63; 96.92%), CIP (n = 62; 95.38%), MEM (n = 60; 92.31%), and CAZ (n = 57; 87.69%) (Figure 1). The imipenem MIC90 for all isolates was ≥16 mg/L. Quite alarmingly, 5 isolates (7.69%) were resistant to all antibiotics tested. Surprisingly, 55 (84.62%) and 35 (53.85%) out of 65 isolates were susceptible to MN and SAM, respectively. Notably, the frequency of MDR isolates was higher in non-ICU wards rather than ICU (Figure 2).
| Isolate No. | Age/Sex | Ward | Outcome | Isolation source | Resistance Patterns | MICIMP | blaOXA23 | blaVIM | blaNDM | intI1 | Class 1 Integron Cassette Arrays | intI2 | Class 2 Integron Cassette Arrays | IntI3 | Clonal Complex |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 42/M | Poisoning ward | ND | S | SAM-MEM-AN-CIP-SXT- CAZ | 8 | + | - | - | + | aadB-aadA1 | - | - | - | CC2 |
| 2 | 63/M | Neurosurgery ICU | Death | T | MEM-AN-CIP-SXT- CAZ | > 64 | - | + | - | + | aadB-aadA1 | - | - | - | CC10 |
| 3 | 50/M | Emergency ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | + | dfrA1-aadA1 | - | - | - | CC2 |
| 4 | 50/F | Emergency ICU | Death | T | MN-SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | + | dfrA1-aadA1 | + | sat2-aadA1 | - | CC2 |
| 5 | 40/M | Poisoning ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | arr2-cm1A5 | + | dfrA1-sat2 | - | CC3 |
| 6 | 41/M | Poisoning ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | dfrA1-aadA1 | - | - | - | CC10 |
| 7 | 75/M | General ICU | Death | T | SAM-AN-CIP-SXT- CAZ | 8 | + | + | - | + | arr2-cm1A5 | - | - | - | CC10 |
| 8 | 50/M | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC10 |
| 9 | 42/M | General ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 8 | - | - | - | + | dfrA1-aadA1 | + | sat2-aadA1 | - | CC3 |
| 10 | 25/F | General ICU | Death | T | MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | - | - | - | - | - | CC10 |
| 11 | 25/F | General ICU | Death | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | dfrA1-aadA1 | - | - | - | CC3 |
| 12 | 37/M | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | - | - | + | dfrA1-sat2 | - | CC3 |
| 13 | 22/M | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | + | arr2-cm1A5 | - | - | - | CC10 |
| 14 | 37/M | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | dfrA1-aadA1 | - | - | - | CC10 |
| 15 | 29/M | Neurosurgery ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 32 | + | - | - | + | arr2-cm1A5 | + | dfrA1-sat2 | - | CC3 |
| 16 | 18/F | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | arr2-cm1A5 | - | - | - | CC10 |
| 17 | 25/F | General ICU | Death | C | MN-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | arr2-cm1A5 | - | - | - | CC2 |
| 18 | 49/M | Neurosurgery ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | + | arr2-cm1A5 | + | dfrA1-sat2 | - | CC2 |
| 19 | 40/F | Neurosurgery ward | Discharge | C | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | - | - | + | sat2-aadA1 | - | CC2 |
| 20 | 96/M | Infectious ward | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | dfrA1-aadA1 | - | - | - | CC2 |
| 21 | 32/F | General ICU | Death | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | arr2-cm1A5 | + | dfrA1-sat2 | - | CC10 |
| 22 | 80/M | Neurosurgery ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | + | dfrA1-aadA1 | - | - | - | CC10 |
| 23 | 5/M | General ICU | Discharge | T | SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC10 |
| 24 | 60/M | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | + | arr2-cm1A5 | - | - | - | CC2 |
| 25 | 32/F | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | arr2-cm1A5 | - | - | - | CC10 |
| 26 | 65/F | Emergency ICU | Death | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | - | - | - | - | - | - | - | CC2 |
| 27 | 75/M | General ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | + | arr2-cm1A5 | + | sat2-aadA1 | - | CC10 |
| 28 | 94/M | Infectious ward | Death | T | MN-SAM-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | aacA4-catB8-aadA1 | + | sat2-aadA1 | - | CC2 |
| 29 | 58/M | Poisoning ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC10 |
| 30 | 33/F | General ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | aadB-aadA1 | - | - | - | CC2 |
| 31 | 29/M | Neurosurgery ICU | Discharge | T | MEM-AN-CIP-SXT | 8 | + | - | - | - | - | - | - | - | CC10 |
| 32 | 80/M | Neurosurgery ICU | Discharge | S | MEM-AN-CIP-SXT- CAZ | >64 | + | + | - | - | - | - | - | - | CC10 |
| 33 | 16/F | Poisoning ICU | Discharge | T | MN-MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | - | - | - | - | - | CC2 |
| 34 | 74/F | General ICU | Death | T | SAM-MEM-AN-CIP-SXT | 8 | + | - | - | + | aacA4-catB8-aadA1 | - | - | - | CC10 |
| 35 | 25/M | Poisoning ICU | Discharge | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | - | - | - | - | - | CC3 |
| 36 | 58/M | Poisoning ICU | Death | T | SAM-AN-SXT | > 64 | + | + | - | + | aadB-aadA1 | + | sat2-aadA1 | - | CC10 |
| 37 | 55/M | Poisoning ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | - | - | - | - | - | CC10 |
| 38 | 74/F | General ICU | Death | T | MEM-AN-CIP-SXT- CAZ | 8 | + | - | - | + | aadB-aadA1 | - | - | - | CC10 |
| 39 | 33/F | General ICU | Death | T | MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | - | - | - | - | - | CC2 |
| 40 | 40/M | Neurosurgery ICU | Discharge | T | MN-MEM-AN-CIP-SXT | 8 | + | - | - | - | - | - | - | - | CC2 |
| 41 | 9/F | General ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | - | - | - | - | - | CC10 |
| 42 | 62/F | General ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | - | - | - | - | - | CC10 |
| 43 | 44/M | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | + | aadB-aadA1 | + | sat2-aadA1 | - | CC10 |
| 44 | 40/F | Poisoning ICU | Death | T | SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | - | - | - | - | - | CC2 |
| 45 | 69/M | General ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | 32 | + | - | - | + | aadB-aadA1 | + | sat2-aadA1 | - | CC2 |
| 46 | 40/F | Poisoning ICU | Death | T | MEM-AN-CIP-SXT | 8 | + | + | - | - | - | - | - | - | CC2 |
| 47 | 69/F | General ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC2 |
| 48 | 80/M | Poisoning ICU | Discharge | T | MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | - | - | - | - | - | CC10 |
| 49 | 41/M | Poisoning ICU | Discharge | T | MN-SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | - | - | - | - | - | CC2 |
| 50 | 92/M | General ICU | Death | T | MN-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | - | - | + | dfrA1-sat2 | - | CC2 |
| 51 | 45/M | Neurosurgery ICU | Discharge | W | MEM-AN-CIP- CAZ | 8 | + | - | - | + | arr2-cm1A5 | - | - | - | CC2 |
| 52 | 50/F | Poisoning ICU | Discharge | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | - | - | - | - | - | CC10 |
| 53 | 55/M | Poisoning ICU | Discharge | T | SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC2 |
| 54 | 45/M | Neurosurgery ICU | Discharge | T | SAM-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | arr2-cm1A5 | - | - | - | CC2 |
| 55 | 69/M | Poisoning ICU | Death | S | MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | + | arr2-cm1A5 | + | dfrA1-sat2 | - | CC10 |
| 56 | 68/F | Neurology ward | Discharge | C | AN-CIP-SXT- CAZ | 32 | - | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC3 |
| 57 | 19/M | Neurosurgery ward | Discharge | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC3 |
| 58 | 65/M | Poisoning ICU | Discharge | T | MEM-AN-CIP- CAZ | 8 | - | - | + | + | aadB-aadA1 | - | - | - | CC10 |
| 59 | 58/M | Neurosurgery ward | Discharge | CF | SAM-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | aadB-aadA1 | - | - | - | CC10 |
| 60 | 46/F | Poisoning ICU | Death | T | MN-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | aacA4-catB8-aadA1 | + | dfrA1-sat2 | - | CC2 |
| 61 | 55/M | Poisoning ICU | Death | T | MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | aadB-aadA1 | - | - | - | CC10 |
| 62 | 58/F | Neurosurgery ICU | ND | T | SAM-MEM-AN-CIP-SXT- CAZ | 16 | + | - | - | + | aadB-aadA1 | - | - | - | CC10 |
| 63 | 17/M | Poisoning ICU | Discharge | T | MN-SAM-MEM-AN-CIP-SXT- CAZ | 32 | + | + | - | + | aacA4-catB8-aadA1 | + | dfrA1-sat2 | - | CC2 |
| 64 | 35/M | Neurosurgery ICU | Discharge | T | CIP-SXT- CAZ | 32 | - | + | - | + | aadB-aadA1 | - | - | - | CC3 |
| 65 | 31/F | General ICU | Death | T | MN-SAM-MEM-AN-CIP-SXT- CAZ | > 64 | + | + | - | + | aacA4-catB8-aadA1 | - | - | - | CC2 |
Abbreviations: SAM, ampicillin/sulbactam; MN, minocycline; MEM, meropenem; AN, amikacin; CIP, ciprofloxacin; SXT, trimethoprim- sulfamethoxazole; CZ, ceftazidime; ND, not determined; S, sputum; T, tracheal; CF, CSF; C, catheter; W, wound.
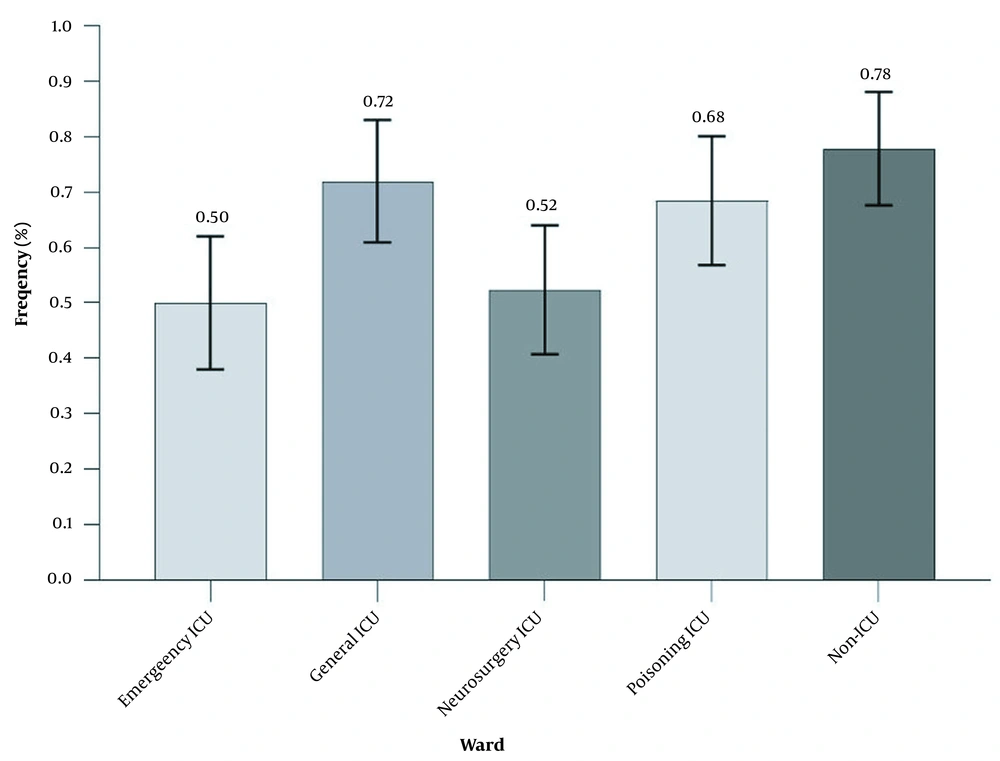
The prevalence of Acinetobacter baumannii isolates obtained from different ICU and non-ICU wards. General ICU had the highest share of clinical samples (72%), while the emergency ICU had the least share of clinical samples (50%). Bars represent mean ± standard deviation (SD). The numbers on the bars represent percentages. ICU, intensive care unit.
4.2. Detection of β-lactamases Encoding Genes
The detected carbapenemase (blaOXA23) and Metallo-β-lactamases (blaVIM and blaNDM) are listed in Table 2. The blaOXA23 was detected in 92.31% (60/65) isolates using PCR amplification. The blaVIM and blaNDM were detected in 69.23% (45/65) and 1.54% (1/65) of the isolates, respectively. Data analysis showed that the presence of blaOXA23 in strains caused a 4- (95% CI, 1.07 - 14.57) and 14-fold (95% CI, 4.22 - 47.50) increase (significantly) in the odds of resistance to SAM and MEM and also the presence of blaVIM caused a 3.5- (95% CI, 1.48 - 8.59) and 11-fold (95% CI, 3.90 - 33.67) increase (significantly) in the odds of resistance to SAM and MEM, respectively.
4.3. PCR Amplification and Characterization of Class 1-3 Integrons
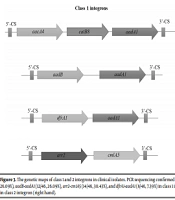
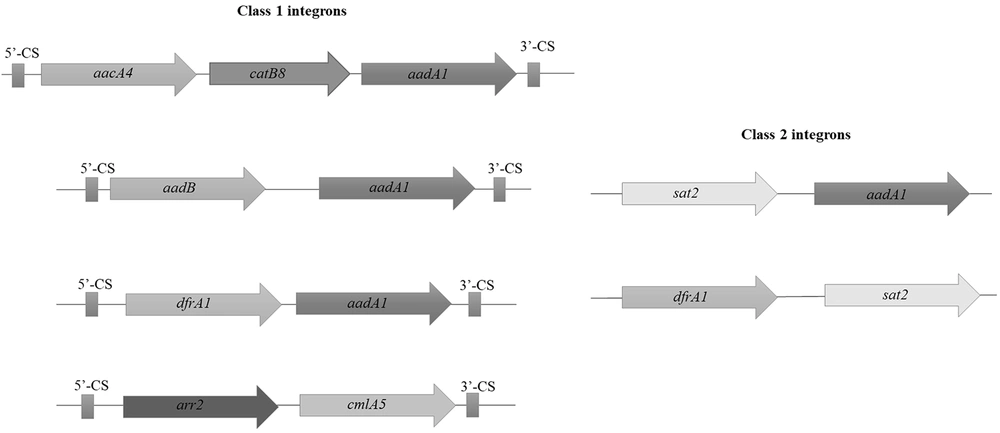
The presence of integrase genes, intI1, intI2, and intI3 were detected by PCR in 70.77% (46/65), 26.15% (17/65), and 0% (0/65) of MDR A. baumannii isolates, respectively. The class 1 and 2 integrons were widespread among clinical isolates. The intI3 was not detected in any of the strains. Cassette arrangements of class 1 and 2 integrons were characterized by PCR sequencing of gene cassettes in the internal variable regions of integrons. Sequencing confirmed the presence of cassette arrays consisting of aacA4-catB8-aadA1 (12/46, 26.09%), aadB-aadA1 (12/46, 26.09%), arr2-cm1A5 (14/46, 30.43%), and dfrA1-aadA1 (8/46, 7.39%) in class 1 integron and dfrA1-sat2 (9/17, 52.94%) and sat2-aadA1 (8/17, 47.06%) in class 2 integron (Figure 3).
The genetic maps of class 1 and 2 integrons in clinical isolates. PCR sequencing confirmed the presence of gene cassette arrays consisting of aacA4-catB8-aadA1 (12/46, 26.09%), aadB-aadA1 (12/46, 26.09%), arr2-cm1A5 (14/46, 30.43%), and dfrA1-aadA1 (8/46, 7.39%) in class 1 integron (left hand) and dfrA1-sat2 (9/17, 52.94%) and sat2-aadA1 (8/17, 47.06%) in class 2 integron (right hand).
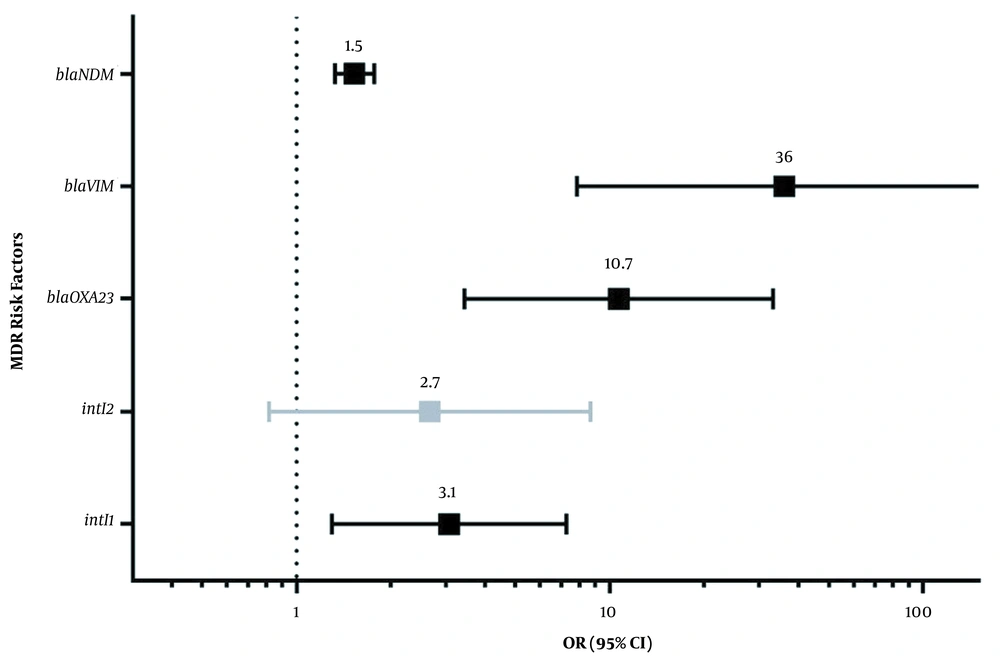
There was no significant difference in resistance to the studied antibiotics among intI1-positive and intI1-negative isolates. The intI1 negative isolates displayed 26.7%, 29.7%, 29%, 30.2%, 24.6%, and 40% resistance rate to SAM, AN, CIP, SXT, CAZ, and MN, respectively, compared with intI1-positive isolates. On the other hand, there was a statistically significant relationship (P < 0.05) between resistance to the tested antibiotics and the lack of the intI2 gene. The intI2-negative isolates displayed 73.3%, 73.4%, 74.2%, 73%, 71.9%, and 50% resistance rate to SAM, AN, CIP, SXT, CAZ, and MN, respectively, compared with intI2 positive isolates. Also, the rate of MDR phenotype in A. baumannii isolates positive for the intI1, blaOXA23, blaVIM, and blaNDM was statistically significant. In addition, although the probability of acquired MDR phenotype for the intI2-positive isolates was 2.7-fold (95% CI, 0.8-8.6) higher than intI2-negative isolates, the latter value was not statistically significant (P > 0.05) (Figure 4).
Odds ratio and 95% confidence interval of risk factors (having the intI1, blaOXA23, blaVIM, blaNDM, or intI2) for acquiring multidrug-resistant phenotypes in Acinetobacter baumannii isolates. The numbers on the bars show the probability of acquired MDR phenotype in each considered risk factor. All of these risk factors were statistically significant except intI2. The dotted vertical line shows a significance threshold.
4.4. Sequence-Based Typing of blaOXA-51-like and ampC Alleles
Sequence-based typing of both blaOXA-51-like and ampC is a discriminatory and reliable method that can distinguish Acinetobacter isolates at the level of clonal complex (23). The SBT results revealed the following distribution of three different clone types among MDR isolates, including CC10 (46.15%, 30/65), CC2 (40%, 26/65), and CC3 (13.85%, 9/65), as shown in Table 2. Overall, 25 out of 30 isolates (83.33%) in CC10, 22 out of 26 isolates (84.62%) in CC2, and 8 out of 9 isolates (88.89%) in CC3 showed MIC ≥16 mg/L for imipenem. Therefore, CC2 and CC10 showed a high level of imipenem resistance. Data analysis showed a heterogenic structure in integron cassette arrays within CCs. The distribution of cassette arrays in class 1 and 2 integrons within clonal complexes is shown in Table 2.
4.5. Nucleotide Accession Numbers
DNA sequences of gene cassette arrays consisting of aacA4-catB8-aadA1 (GenBank accession number = MZ508285), aadB-aadA1 (MZ508283), arr2-cm1A5 (MZ508286), and dfrA1-aadA1 (MZ508284), in class 1 integron and dfrA1-sat2 (MZ508287) and sat2-aadA1 (MZ508288) in class 2 integron were deposited in GenBank database.
5. Discussion
Infections associated with MDR bacterial strains have become one of the leading causes of morbidity and mortality worldwide (24). Integrons as transposon-like genetic elements are conserved and encode antibiotic resistance determinants and have a high capacity for chromosomal integration in bacteria (25, 26). To date, several classes of integrons have been described, of which class 1 and 2 integrons are commonly reported from MDR A. baumannii strains (27). Carbapenems are usually the antibiotic of choice against A. baumannii strains. However, the rate of resistance to carbapenems in this bacterium is increasing day by day. Resistance to carbapenems can be due to various mechanisms, such as producing the enzymes, including Metallo-β-lactamase and oxacillinase (28, 29).
According to our results, most MDR A. baumannii isolates were obtained from the tracheal aspirate samples. Consistent with our research, in a study conducted by Souza et al., A. baumannii was the most frequently isolated bacterial species in the tracheal secretion of patients with ventilator-associated pneumonia (30). However, Barbier et al. reported that the most frequent pathogens associated with ventilator-associated pneumonia were Staphylococcus aureus, Pseudomonas aeruginosa, and Enterobacteriaceae (31). The ICU has been described as the main center of antibiotic resistance development, with increasingly resistant isolates complicating the treatment of MDR infections in ICU patients (32). Surprisingly, the frequency of MDR A. baumannii isolates was higher in non-ICU wards rather than ICU. This is alarming and indicates that MDR isolates have been circulated in our hospital, and urgent attention and application of preventive protocols are needed to reduce such a fearful threat in hospitalized patients. In this study, the highest antibiotic resistance was related to amikacin, trimethoprim + sulfamethoxazole, and ciprofloxacin, respectively. Also, A. baumannii strains producing aminoglycoside modifying enzymes (AMEs) are highly resistant to different aminoglycosides, such as gentamicin, amikacin, and tobramycin. Similar to our findings, Cho et al. reported aminoglycoside resistance genes in 81% Acinetobacter isolates from two Korean hospitals (33).
In this study, the imipenem MIC90 for all isolates was ≥ 16 mg/L and showed resistance to carbapenems. According to a study conducted by Lee et al., isolates with MIC ≤ 4 mg/L were susceptible to carbapenem, and those with MIC ≥ 8 mg/L were resistant in patients with A. baumannii bacteremia (34). Consistent with our results, Akbari Dehbalaei et al. reported that resistant to carbapenems was up to 85% in A. baumannii isolates (35). Unfortunately, in this study, 7.69% of the isolates were resistance to all tested antibiotics, which will be a significant obstacle to effective treatment in the future. Therefore, antibiotic usage should be controlled to prevent this serious threat. On the other hand, 84.62% and 53.85% of isolates were susceptible to MN and SAM, respectively. This result indicated that these two antibiotics could be effective for the treatment of A. baumannii infections in combination form. However, excessive usage of these two antibiotics can also increase antibiotic resistance against them.
In this study, blaOXA-23, blaVIM, and blaNDM were detected with high frequency in A. baumannii isolates. The blaOXA-23-like gene is one of the most prevalent β-lactamase genes in the carbapenem-resistant A. baumannii genome, mostly on plasmids (36). Specific and quick identification of A. baumannii and strains containing the blaOXA-23-like gene will reference information on treatment and control measures for carbapenem resistance (37). Ning et al. showed that ST191 and ST195 isolates of OXA-23-producing A. baumannii could spread in a hospital and became potential nosocomial outbreak strains. In this regard, they suggested that antimicrobial management and surveillance of imipenem-resistant A. baumannii should be improved (38). Moreover, in the study by Akbari Dehbalaei et al., the blaOXA-23 gene was detected in 81.81% of the isolates.
This study concluded that highly resistant blaOXA-23 gene-harboring endemic clones of A. baumannii were disseminated in the ICUs of two studied hospitals (35). The blaVIM is another β-lactamases encoding genes with a frequency of 69.23% in this study. The frequency of this gene was reported to be 17.44% and 18.18% in other studies conducted in Iran in 2014 and 2016, respectively (39, 40). Comparison of these results showed that the frequency of this gene had increased significantly in recent years in Iran (39, 40). Therefore, it seems necessary to find new treatments to deal with this problem. In this study, only one isolate harbored the blaNDM-1 gene. Pillonetto et al. presented the first instance of A. baumannii sequence type 25 generating blaNDM-1, isolated from the urinary tract of a 71-year-old man in Brazil (41). Bonnin et al. recently suggested that A. baumannii may accept resistant genes and act as a gene donor passing resistance genes to other bacteria, including Enterobacteriaceae (42). It seems that the MDR phenotype in A. baumannii is associated with the cooperation of carbapenemases, class 1 integrons, and possibly efflux pumps.
The presence of integrase genes, intI1 and intI2, was detected by PCR in 70.77% and 26.15% of A. baumannii isolates, respectively. These data indicated that class 1 and 2 integrons were widely distributed among clinical isolates of A. baumannii. The intI3 was not detected in any of the strains. Similar to our study, Goudarzi and Azimi reported class 1 and 2 integrons in 66.7% and 20% of isolates, respectively. However, the class 3 integron was detected in three A. baumannii strains (8). Moreover, Nourbakhsh et al. reported the frequency of class 1, 2, and 3 integrons to be 100%, 44%, and 3%, respectively, among A. baumannii isolates (43). The sequence-based typing results of blaOXA-51-like and ampC alleles revealed the following distribution of three different clone types among MDR isolates, including CC10 (46.15%), CC2 (40%), and CC3 (13.85%). In the study by Nazari et al., a comparison of clonal relatedness between clinical and non-clinical isolates illustrated that widespread clones, including CC2, CC3, and CC10 were common clonal complexes among clinical and non-clinical strains (15). In addition, a systematic review on clonal relatedness of A. baumannii isolated from the Middle East showed that CC2 was the most prevalent clonal complex isolated from Lebanon, Palestine, Saudi Arabia, Turkey, Yemen, Iran, Iraq, and Kuwait. In this study, CC2 and CC10 showed a high-level imipenem resistance (44).
5.1. Conclusions
The high prevalence of carbapenemase-producing A. baumannii isolates in the ICU requires a rigorous antimicrobial stewardship and infection control program. Class 1 and 2 integrons in clinical strains are repertoires of aminoglycoside-modifying enzymes. Class 1 integron can be served as a predictive biomarker for the presence of MDR bacteria in the clinical setting. However, hoarding of carbapenemases on the integron apparatus is not widespread among A. baumannii strains. Continuous surveillance MDR A. baumannii and elucidation of their AMR mechanisms in the clinical setting are clearly necessary to help develop effective therapy regimens and to prevent the further dissemination of these superbug bacteria. Further studies are required to elaborate the association of gene pools in A. baumannii and antibiotic resistance patterns with epidemic and clinical outcomes of infection.