1. Background
Enterococcus faecalis is the normal flora of the gastrointestinal system of humans and animals. As an occasional pathogen, it can cause community-acquired and nosocomial infections (1). It is responsible for different nosocomial infections, particularly bacteremia, sepsis, endocarditis, wound infections, and urinary tract infections (2). Previous studies have revealed that most enterococcal infections are caused by E. faecalis strains (3, 4). Enterococcal infections are a major challenge to healthcare systems and a serious threat to patient life in Iran (5). In Iran, a study on 111 clinical Enterococcus isolates showed that 80.1% of the isolates were E. faecalis (6). Hospitalization and impaired immunity are the risk factors of E. faecalis infections (7). Moreover, antimicrobial resistance and virulence factors remarkably contribute to enterococcal infections in hospitalized patients (8). In addition to innate resistance to several antimicrobial agents, Enterococcus species can acquire high resistance to different antibiotics through the horizontal transfer of mobile genetic elements (9).
Given the wide and inappropriate use of antibiotics in clinical settings for managing community-acquired infections, the emergence of multidrug-resistant (MDR) E. faecalis strains has become a serious healthcare concern worldwide. There are limited treatment options for MDR bacterial species. Moreover, MDR species can transfer genetic antimicrobial resistance factors to other commensal bacteria in the gastrointestinal system or the environment (10). Accordingly, there are clinical challenges in managing enterococcal infections due to their progressive resistance to different antibiotics such as beta-lactams, macrolides, fluoroquinolones, glycopeptides, and aminoglycosides (11, 12).
Antimicrobial resistance to the high concentrations of aminoglycoside antibiotics is usually caused by the aminoglycoside modifying enzymes found in and encoded by mobile genetic elements. Moreover, resistance to vancomycin is encoded by the van gene clusters transferred in one transposon (13). Virulence factors such as gelatinase, aggregation substance proteins, Enterococcus surface proteins, collagen adhesion, cytolysin, and hyaluronidase play roles in bacterial adhesion, colonization, invasion, escaping from the host’s immune response, extracellular enzyme production, virulence, and infection aggravation (14).
Source detection and the effective management of enterococcal nosocomial infections largely depend on determining their genetic diversity (15). Currently, different methods such as pulse-field gel electrophoresis (PFGE), restriction fragment length polymorphism (RFLP), and enterobacterial repetitive intergenic consensus using polymerase chain reaction (ERIC-PCR) have been used for the epidemiological assessment of bacteria (16, 17). ERIC-PCR is a popular molecular analysis method for typing Enterococcus isolates. The ERIC sequences are the imperfect palindromes of 127 base pairs (bp); however, the ERIC region can be smaller or larger due to internal eliminations or the entrance of larger sequences. The ERIC sequences were first detected in Escherichia coli, Salmonella typhimurium, other Enterobacteriaceae members, and Vibrio cholerae. However, it has recently been revealed that they are present in other species and can be used for typing (18). In Indonesia, a study showed that all E. faecalis isolates analyzed by the Eric typing method had >60% similarity and were in five clusters (19).
Several studies in different countries reported the great antimicrobial resistance and the great genetic diversity of Enterococcus species (20-22). A study on Enterococcus isolates obtained from the stool samples of children in the northwest of Iran showed great antimicrobial resistance to tetracycline, rifampin, and erythromycin and great genetic diversity among the E. faecalis isolates (23). The genetic pattern of Enterococcus species such as E. faecalis varies not only in different areas but also in a single setting; hence, there may be multiple clusters specific to a single area or setting.
2. Objectives
Studying the epidemiology of E. faecalis can provide valuable information about the current and future status of nosocomial infections. The present study aimed to analyze the genetic diversity, antimicrobial resistance, and virulence factors of E. faecalis isolates obtained from the stool samples of patients in a hospital in the center of Iran.
3. Methods
3.1. Sampling and Enterococcus faecalis Detection
This cross-sectional descriptive-analytical study was conducted from September 2019 to February 2020. The non-duplicated stool samples were obtained from patients hospitalized for at least three days in the internal medicine ward, infectious diseases ward, and coronary care unit of the Shahid Beheshti Hospital, Kashan, Iran. The isolates were cultured in the laboratory of the Microbiology Department of the Kashan University of Medical Sciences, Kashan, Iran. They were phenotypically detected at the species level using biochemical tests such as catalase test, culturing on the brain heart infusion broth containing 6.5% NaCl, and the Bile-Esculin test (24). The identity of the isolates was genotypically confirmed using the PCR method and the specific primer of the ddlE gene of E. faecalis (25).
3.2. Antimicrobial Resistance Testing
The antimicrobial resistance pattern of E. faecalis isolates was determined phenotypically and genotypically. In the phenotypic method, the disc agar diffusion method was used according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) to test the antimicrobial resistance of the isolates for the following 10 antibiotics: penicillin (PEN, 10 units), ampicillin (AMP, 10 µg), ciprofloxacin (CIP, 5 µg), erythromycin (E, 15 µg), tetracycline (TET, 30 µg), nitrofurantoin (NI, 300 µg), rifampin (RI, 5 µg), quinupristin-dalfopristin (SYN, 15 µg), and linezolid (LZD, 5 µg). First, a 0.5 McFarland microbial suspension of the isolates was prepared. After culturing the isolates, the inhibition zone diameter was measured and interpreted based on the CLSI table (26). According to the recommendations of the CLSI, Staphylococcus aureus ATCC 25923 was used as a control to standardize the antimicrobial resistance test. Isolates resistant to at least three antibiotic classes were considered as MDR.
3.3. DNA Extraction
DNA extraction was performed using the DNA TIANamp extraction kit (TIANGEN Biotech, Beijing, China). The spin column in this kit is made of a silica membrane, which can attach to and detach from DNA under specific pH and salt conditions. The DNA concentration was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA).
3.4. Detection of the Antimicrobial Resistance and the Virulence Genes
The PCR method was employed to genotypically detect the genes responsible for resistance to antibiotics vancomycin (vanB, vanA), aminoglycosides (aac(6′)-Ie aph(2′′)Ia), and macrolides (ermA, ermB, ermC) (Table 1). To this end, specific primers and the method described in our previous work (27) were used. Moreover, E. faecalis isolates were assessed for the virulence genes efa, ace, asa1, gelE, hy1, cy1A, and esp using specific primers and PCR conditions, as explained in Table 2.
| Target Gene | Primers | Oligonucleotide Sequence 5'-3' | Size of Amplified Product (bp) | Annealing Temperature |
|---|---|---|---|---|
| vanA | F | GGGAAAACGACAATT GC | 732 | 54°C |
| R | GTA CAATGC GGCCGTTA | |||
| vanB | F | ATGGGAAGCCGATAGTC | 638 | |
| R | GATTTCGTTCCTCGACC | |||
| aac(6′)-Ie | F | AGGAATTTATCGAAAATGGTAGAAAAG | 369 | 54°C |
| R | CACAATCGACTAAAGAGTACCAATC | |||
| aph(2′′)Ia | F | CAAACTGCTAAATCGGTAGAAGCC | 294 | |
| R | GGAAAGTTGACCAGACATTACGAACT | |||
| ermA | F | TAT CTT ATC GTT GAG AAG GGA TT | 139 | 55°C |
| R | CTA CAC TTG GCT GAT GAA A | |||
| ermB | F | CTA TCT GAT TGT TGA AGA AGG ATT | 142 | |
| R | GTT TAC TCT TGG TTT AGG ATG AAA | |||
| ermC | F | AAT CGT CAA TTC CTG CAT GT | 299 | |
| R | TAA TCG TGG AAT ACG GGT TTG |
| Target Gene | Primers | Oligonucleotide Sequence 5'-3' | Size of Amplified Product (bp) | Annealing Temperature |
|---|---|---|---|---|
| asa1 | F | GCACGCTATTACGAACTATGA | 375 | 56°C |
| R | TAAGAAAGAACATCACCACGA | |||
| esp | F | AGATTTCATCTTTGATTCTTG | 510 | 56°C |
| R | AATTGATTCTTTAGCATCTGG | |||
| hyl | F | ACAGAAGAGCTGCAGGAAATG | 276 | 56°C |
| R | GACTGACGTCCAAGTTTCCAA | |||
| gelE | F | TATGACAATGCTTTTTGGGAT | 213 | 56°C |
| R | AGATGCACCCGAAATAATATA | |||
| cylA | F | GACTCGGGGATTGATAGGC | 688 | 56°C |
| R | GCTGCTAAAGCTGCGCTTAC | |||
| ace | F | GGAATGACCGAGAACGATGGC | 616 | 58°C |
| R | GCTTGATGTTGGCCTGCTTCCG | |||
| efaA | F | CGTGAGAAAGAAATGGAGGA | 499 | 58°C |
| R | CTACTAACACGTCACGAATG |
3.5. ERIC Typing
Genetic diversity was assessed using ERIC-PCR, as described by Versalovic et al. (28). The ERIC region was detected using the ERIC1R (5'-ATG TAA GCT CCT GGG GAT TCAC-3') and the ERIC2 (5'-AAG TAA GTG ACT GGG GTG AGC G-3') primers (Metabion, Germany). Moreover, the ERIC region was amplified using a 25-µL mixture of sterile distilled water (18 µL), 10X PCR buffer (2.5 µL), dNTP (1 µL of 10 molars), ERIC1R and ERIC2 primers (1 µL of each), Taq polymerase (0.5 µL), and template DNA (1 µL). The PCR reaction consisted of initial denaturation at 95°C for three minutes accompanied with 35 warming cycles, each of which consisted of denaturation at 94°C for one minute, annealing at 48°C for one minute, extension at 72°C for two minutes, and final extension at 72°C for five minutes (29).
3.6. ERIC Analysis
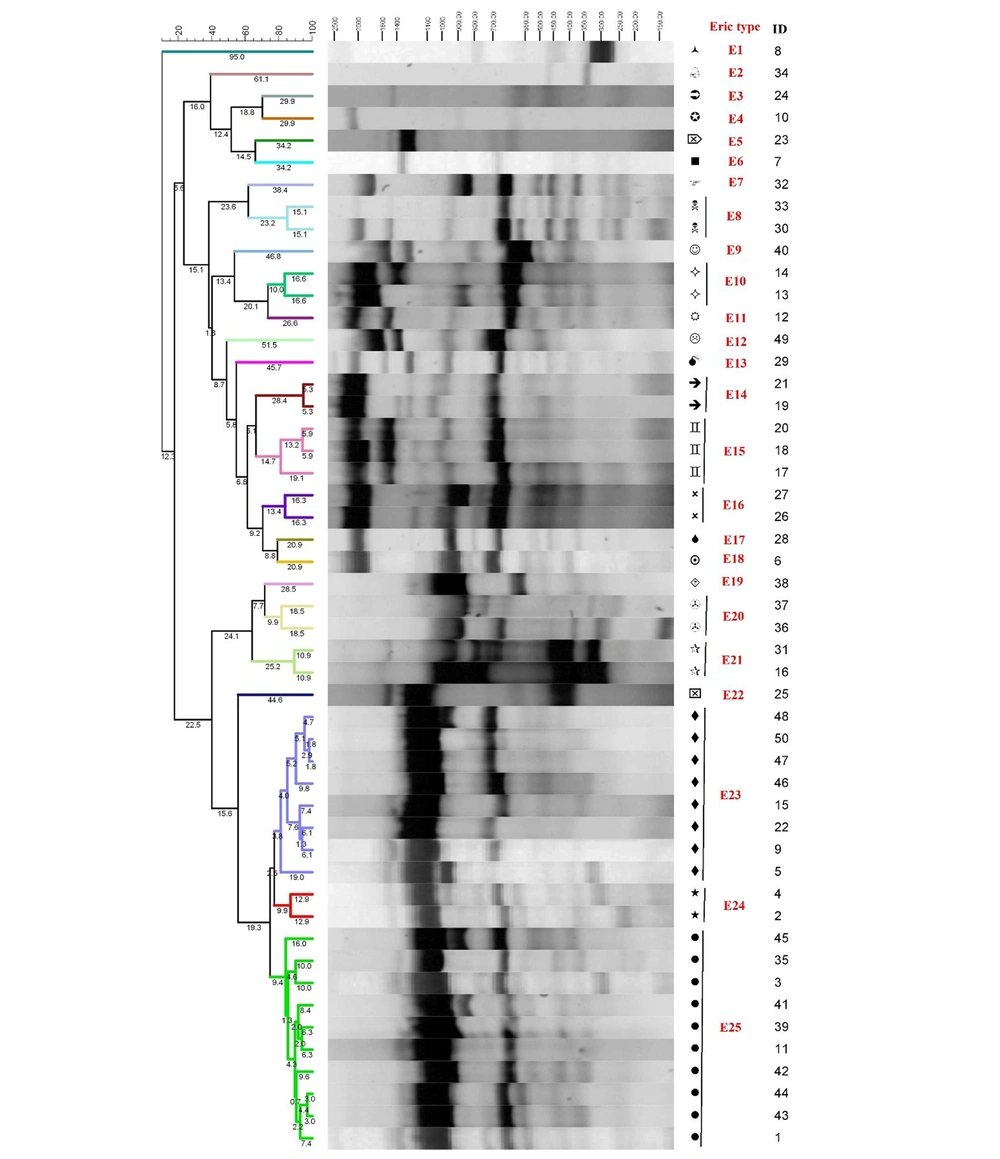
The band profile analysis was done with BioNumerics software version 8.0. The dendrogram was constructed by the genetic diversity analysis using the Unweighted Pair Group Mean Method with Arithmetic mean (UPGMA), disc similarity coefficient, and 1% band position tolerance. According to the ladder, only the bands with a length of 100 - 3000 bp were included in the analysis. The BioNumerics software creates groups with a specific similarity range. The differences between band profiles in the dendrogram were depicted using the digits above lines (Figure 1). For the band analysis, the isolates with a genetic similarity of at least 80% were grouped as an ERIC type; however, the isolates not having such a similarity were considered as separate ERIC types and then labeled as ERIC E1–En. ERIC clustering criteria in the BioNumerics software were a difference of ≤ 20% and the presence of more than one isolate in each cluster.
3.7. Statistical Analysis
The collected data was analyzed with SPSS software version 22.0. Values were reported as relative frequency. The chi-square test was used for frequency comparison at P < 0.05.
4. Results
4.1. Characteristics of Patients and Isolates
The phenotypic and genotypic tests were used, and fifty E. faecalis isolates were obtained from 108 stool samples (46.2%). None of the samples was duplicated. The E. faecalis isolates were obtained from an internal medicine ward (n = 22), an infectious diseases ward (n = 16), and a coronary care unit (n = 12). The Enterococcus faecalis isolates were obtained from both male (n = 29) and female (n = 21) patients. The participants’ mean age was 65.1 ± 15.4 years (Table 1).
4.2. Antimicrobial Resistance Testing
The antimicrobial resistance testing results showed that the great resistance of the E. faecalis isolates to the antibiotics TET (70%), E (68%), RI (60%), SYN (48%), and CIP (48%). Table 3 shows the antimicrobial resistance of E. faecalis isolates to the ten tested antibiotics. Thirty-one E. faecalis isolates (62%) were MDR. In this regard, MDR had no significant relationship with patients’ gender, age, length of hospital stay, and hospitalization ward (P > 0.05). The Enterococcus faecalis isolates showed the highest antimicrobial susceptibility to the antibiotics LZD (94%), NI (86%), AMP (78%), and PEN (76%).
| No. | Gender | Age (y) | Ward | Hospital Stay (d) | Resistance Patterns | Resistance Genes | MDR | Virulence Genes | ERIC Type | Cluster |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | 60 | IM | 5 | _ | ermB, ermC, aac(6′)-Ie | asa1, esp, gelE | E25 | J | |
| 2 | Female | 84 | CCU | 4 | E+SYN+TET | ermB, aph(2′′)Ia | + | asa1, esp | E24 | I |
| 3 | Female | 63 | IM | 7 | RI+E+SYN+PEN+CIP | ermB, aph(2′′)Ia | + | asa1, esp, gelE | E25 | J |
| 4 | Female | 56 | IM | 3 | E+SYN+TET +CIP | ermB, aac(6′)-Ie, aph(2′′)Ia | + | asa1, esp, gelE | E24 | I |
| 5 | Male | 65 | IM | 6 | RI+E+SYN+CIP | ermB, ermC, aph(2′′)Ia | + | asa1, gelE | E23 | H |
| 6 | Female | 70 | IM | 3 | RI+E+PEN+TET+CIP+AMP | vanA, ermB | + | asa1, esp | E18 | — |
| 7 | Female | 75 | IM | 6 | E+SYN | ermB | gelE | E6 | — | |
| 8 | Male | 82 | CCU | 4 | RI+SYN+CIP | ermB, aac(6′)-Ie | + | — | E1 | — |
| 9 | Male | 74 | IM | 5 | RI+E+SYN+PEN+TET+CIP+AMP | ermB, aac(6′)-Ie, aph(2′′)Ia | + | asa1, gelE | E23 | H |
| 10 | Female | 78 | CCU | 3 | SYN+TET | ermB, ermC, aph(2′′)Ia | cylA, asa1, esp, gelE | E4 | — | |
| 11 | Female | 66 | IM | 3 | RI+E+SYN+TET+CIP | ermA, ermB, aac(6′)-Ie, aph(2′′)Ia | + | asa1, esp, gelE | E25 | J |
| 12 | Male | 55 | IM | 3 | _ | ermB, ermC, aac(6′)-Ie, aph(2′′)Ia | esp | E11 | — | |
| 13 | Female | 17 | CCU | 3 | TET | vanA, ermB, aac(6′)-Ie, aph(2′′)Ia | cylA, asa1, esp, gelE | E10 | B | |
| 14 | Female | 67 | CCU | 3 | E+NI+TET | ermB | + | asa1, esp | E10 | B |
| 15 | Male | 48 | CCU | 3 | RI+E+SYN+TET | ermB | + | asa1, esp, gelE | E23 | H |
| 16 | Male | 86 | CCU | 7 | E+TET | ermB, aph(2′′)Ia | esp, gelE | E21 | G | |
| 17 | Male | 83 | CCU | 3 | TET | ermB | cylA, asa1, esp, gelE | E15 | D | |
| 18 | Female | 80 | IM | 4 | _ | ermB | gelE | E15 | D | |
| 19 | Female | 59 | IM | 3 | RI+E+PEN+TET+CIP+AMP | vanA, ermB, aph(2′′)Ia | + | cylA, asa1, esp, gelE | E14 | C |
| 20 | Female | 59 | IM | 4 | _ | ermB | cylA, asa1 | E15 | D | |
| 21 | Female | 79 | CCU | 4 | _ | vanA, ermB, aph(2′′)Ia | cylA, asa1, esp, gelE | E14 | C | |
| 22 | Female | 86 | IM | 5 | TET | ermB, aph(2′′)Ia | gelE | E23 | H | |
| 23 | Male | 75 | ID | 4 | E+CIP | ermB, aph(2′′)Ia | asa1, esp, gelE | E5 | — | |
| 24 | Male | 41 | IM | 3 | E+TET | ermB | cylA, asa1, esp | E3 | — | |
| 25 | Female | 54 | IM | 3 | RI+E+PEN+TET+CIP+AMP | ermB, ermC, aac(6′)-Ie | + | asa1, esp, gelE | E22 | — |
| 26 | Male | 42 | IM | 3 | TET | _ | cylA, asa1, esp, gelE | E16 | E | |
| 27 | Female | 78 | IM | 5 | E+CIP | ermB, aph(2′′)Ia | asa1, esp | E16 | E | |
| 28 | Female | 45 | ID | 6 | RI+E+CIP | ermB, aph(2′′)Ia | + | cylA, asa1, esp, gelE | E17 | — |
| 29 | Female | 77 | IM | 4 | RI+E+TET | ermB, aph(2′′)Ia | + | cylA, asa1, esp, gelE | E13 | — |
| 30 | Male | 44 | ID | 8 | RI+E+SYN+TET+CIP | ermB, aph(2′′)Ia | + | asa1, esp, gelE | E8 | A |
| 31 | Female | 43 | ID | 10 | RI+TET | ermA, ermB, ermC, aph(2′′)Ia | asa1, esp, gelE | E21 | G | |
| 32 | Male | 51 | ID | 6 | RI+SYN | ermB, ermC, aac(6′)-Ie, aph(2′′)Ia | asa1, esp, gelE | E7 | A | |
| 33 | Male | 53 | CCU | 5 | RI+E+SYN+NI+PEN+TET+CIP+AMP | vanA, ermB, ermC aac(6′)-Ie, aph(2′′)Ia | + | asa1, esp, gelE, hyl | E8 | — |
| 34 | Male | 73 | ID | 7 | RI+E+LZD+NI+PEN+TET+CIP+AMP | ermB, aac(6′)Ie, aph(2′′)Ia | + | asa1, esp, gelE | E2 | — |
| 35 | Male | 54 | ID | 4 | RI+SYN+TET | ermB, ermC, aac(6′)-Ie, aph(2′′)Ia | + | asa1, esp | E25 | J |
| 36 | Male | 68 | ID | 5 | RI+E | ermB, aac(6′)Ie, aph(2′′)Ia | asa1, esp, gelE | E20 | F | |
| 37 | Male | 49 | ID | 4 | RI+E+SYN+TET | ermA, ermB, ermC, aac(6′)Ie | + | asa1, esp, gelE | E20 | F |
| 38 | Male | 80 | ID | 3 | RI+E+SYN+NI+PEN+TET+CIP+AMP | vanA, ermA, ermB, ermC, aac(6′)Ie, aph(2′′)Ia | + | esp, gelE | E19 | — |
| 39 | Male | 71 | ID | 3 | RI+E+SYN+NI+PEN+TET+CIP+AMP | vanA, ermB, ermC, aac(6′)Ie, aph(2′′)Ia | + | asa1, esp, gelE | E25 | J |
| 40 | Male | 65 | ID | 3 | RI+E+SYN+TET | ermB, aac(6′)Ie | + | asa1, esp, gelE | E9 | — |
| 41 | Male | 90 | IM | 5 | RI+NI+TET+CIP | ermB | + | — | E25 | J |
| 42 | Male | 52 | ID | 7 | RI+TET | ermA, ermB, ermC, aac(6′)Ie, aph(2′′)Ia | asa1, esp, gelE | E25 | J | |
| 43 | Male | 71 | ID | 3 | RI+E+ PEN+CIP+AMP | vanA, ermA, ermB, ermC, aac(6′)Ie, aph(2′′)Ia | + | asa1, esp, gelE | E25 | J |
| 44 | Male | 89 | ID | 3 | RI+E+SYN+PEN+TET+CIP+AMP | ermB, aac(6′)Ie, aph(2′′)Ia | + | asa1, esp, gelE | E25 | J |
| 45 | Male | 65 | IM | 6 | E+SYN+ TET+CIP | ermB, aph(2′′)Ia | + | asa1, gelE | E25 | J |
| 46 | Male | 80 | IM | 3 | RI+E+SYN+ TET+CIP | vanA, ermB, aph(2′′)Ia | + | esp, gelE | E23 | H |
| 47 | Male | 45 | ID | 4 | RI+E+SYN+PEN+TET+CIP+AMP | ermA, ermB, ermC, aac(6′)Ie, aph(2′′)Ia | + | _ | E23 | H |
| 48 | Male | 73 | IM | 4 | RI+E+LZD+SYN+NI+TET | ermB | + | gelE | E23 | H |
| 49 | Female | 63 | CCU | 3 | E+SYN+TET+CIP | vanA, ermB, aph(2′′)Ia | + | gelE | E12 | — |
| 50 | Male | 73 | CCU | 3 | RI+E+LZD+TET | _ | + | gelE | E23 | H |
Abbreviations: IM, internal medicine; CCU, coronary care unit; ID, infectious disease.
4.3. Antimicrobial Resistance Genes
The analysis of antimicrobial resistance genes among the E. faecalis isolates indicated that the most prevalent genes were ermB (96%), aph(2′′)Ia (66%), aac(6′)-Ie (40%), ermC (30%), vanA (20%), and ermA (14%). Moreover, the most prevalent antimicrobial resistance patterns were ermB+ aph(2′′)Ia (20%), ermB (18%), and ermB+aac(6′)-Ie+ aph(2′′)Ia (10%).
4.4. Virulence Genes
The virulence gene analysis of the E. faecalis isolates showed that the most prevalent genes were gelE (78%), asa1 (74%), and esp (74%), and that the prevalence rates of the cylA and hyl genes were low (20% and 2%, respectively). Moreover, the most prevalent virulence gene patterns were asa1+esp+gelE (36%) and cylA+asa1+esp+gelE (16%).
4.5. Genetic Diversity Analysis
The genetic diversity analysis by ERIC-PCR revealed 25 genotypes among fifty E. faecalis isolates (E1–E25), which consisted of fifteen genotypes with a genetic similarity of at least 80% and ten genotypes with a genetic similarity of < 80%. Each of the fifteen genotypes was placed in an ERIC type (Figure 1). The dendrogram analysis revealed that most E. faecalis isolates (n = 35) were clustered into ten clusters (ie, clusters A–J). Eighteen isolates were further clustered into two major clusters: cluster H with eight isolates and cluster J with ten isolates. The remaining seventeen isolates were clustered into eight minor clusters: A, B, C, D, E, F, G, and I, each with 2 - 3 isolates (Figure 1).
The antimicrobial resistance analysis in the two major clusters H and J revealed the greatest resistance to the antibiotics E, RI, and TET. Moreover, the antimicrobial resistance gene analysis also revealed that the most prevalent genes in cluster H were ermB (87%) and aph(2 ′′)Ia (62.5%). In comparison, the most prevalent genes in cluster J were ermB (100%), aph(2′′)Ia (80%), and aac(6′)-Ie (70%). However, Fisher's exact test indicated no significant difference between clusters H and J in terms of antimicrobial resistance and resistance genes. The virulence gene analysis also suggested that the most prevalent genes in cluster H were gelE (100%) and asa1 (37.5%), and the most prevalent genes in cluster J were asa1 (90%), gelE (80%), and esp (80%). The prevalence of the asa1 gene was significantly higher in cluster J than in cluster H (P < 0.05).
5. Discussion
This study analyzed the genetic diversity, antimicrobial resistance, and virulence factors of E. faecalis isolates obtained from the stool samples of patients in a hospital in the center of Iran. In this study, fifty E. faecalis isolates were obtained from 108 stool samples. In other words, the prevalence of E. faecalis isolates was 46.2%. This prevalence rate exceeds the rate reported in a study in China (15%) and is below the rate reported in a study in Brazil (70%) (30). Several recent studies reported that the prevalence of E. faecalis isolates among clinical Enterococcus isolates was 28 - 70% (5, 31, 32). This difference in the prevalence of E. faecalis isolates in these studies can be due to the differences among clinical settings regarding their infection control policies.
The findings showed that the 62-percent prevalence rate of MDR patterns among the E. faecalis isolates. Antimicrobial resistance among enterococcal serotypes is a major public health concern worldwide (33). A study in Iran showed that 29% of the E. faecalis isolates obtained from urinary tract infections were MDR (34). Another study at the Shahid Beheshti Hospital, Kashan, Iran, reported that the prevalence rate of 37.7% for the MDR E. faecalis isolates obtained from clinical samples (35). The higher prevalence of MDR among the E. faecalis isolates in our study denotes the high risk of enterococcal nosocomial infections in the study setting. The MDR isolates cause treatment failure and are associated with a higher mortality rate compared wo the non-MDR isolates (36). Factors such as healthcare exposure, shortage of diagnostic equipment, lack of surveillance systems, and the wide and inappropriate use of antibiotics can contribute to the emergence and spread of antimicrobial resistance, the acquisition of antimicrobial resistance genes, and the transfer of these genes. These factors might also have resulted in the high prevalence of antimicrobial resistance in the present study.
The research findings also revealed great antimicrobial resistance to antibiotics TET (70%), E (68%), and RI (63%) among the E. faecalis isolates obtained from the stool samples of hospitalized patients. This is in line with the findings of previous studies in Iran (37), Greece (38), and China (39). However, studies in India (40) and Brazil (41) reported lower antimicrobial resistance to these antibiotics. The use of over-the-counter antibiotics for infection management and agriculture may be the reasons for great resistance to antibiotics such as TET in Iran. Most E. faecalis isolates in the present study were susceptible to LZD. In Spain, a study showed that 100% of the E. faecalis isolates were susceptible to LZD and AMP (42). Another study in our setting also reported that the antimicrobial susceptibility of the E. faecalis isolates to antibiotics LZD and AMP was 100% and 88%, respectively (35). Nonetheless, our findings showed three LZD-resistant isolates, and the susceptibility of the E. faecalis isolates was 94% to LZD and 78% to AMP, which are slightly smaller than the rates reported in the aforementioned study.
The antimicrobial resistance gene analysis in the present study showed that the most prevalent genes were ermB (96%), aph(2′′)Ia (66%), aac(6′)-Ie (40%), ermC (30%), vanA (20%), and ermA (14%). Genetic antimicrobial resistance attributes are either innate or acquired and can be transferred to other isolates (10). A mechanism for cross-resistance to macrolide-lincosamide-streptogramin among E. faecalis isolates is to change the target site of the antibiotic erythromycin, which is mediated by the ermB gene (43). The high prevalence of this gene in the present study is in line with the findings of studies in the United States, China, South Korea, and Saudi Arabia (43). However, the prevalence of the ermA gene in the present study was 14%.
This finding is in contrast with the findings of former studies, which reported that above 90% of E. faecalis isolates had the ermA gene (44, 45). The prevalence rates of the aph(2′′)Ia and aac(6′)-Ie genes in the present study were 66% and 40%, respectively. These genes encode the 6-aminoglycoside acetyltransferase and 2-aminoglycoside phosphotransferase enzymes found in most aminoglycoside-resistant Enterococcus species. The incidence of these aminoglycoside-resistant genes has been increased in recent years (46). Enterococcus species with these genes have great antimicrobial resistance; hence, they, can escape antimicrobial mechanisms and cause infection in their hosts.
The prevalence rates of the vanA and the vanB genes in the E. faecalis isolates were 20% and 0%, respectively. The emergence of vancomycin-resistant Enterococcus species and the limited treatment options for their management have become a major clinical and epidemiological concern since afflicted patients experience more complications and higher treatment costs. Nine types of cluster genes encode vancomycin resistance, among which vanA–vanN. vanA and vanB are the most important and prevalent genes in vancomycin-resistant Enterococcus species (45). Genetic factors such as plasmids, integrons, and transposons contribute to the emergence, transfer and the spread of antimicrobial resistance genes, particularly in Enterococcus species, thereby resulting in treatment failure (47).
The virulence gene analysis in the present study also revealed that the most prevalent virulence genes were gelE (78%), asa1 (74%), and esp (74%), followed by the less prevalent genes of cylA (20%) and hyl (2%). These findings documented that most E. faecalis isolates carried the gelE, asa1, and esp genes. A previous study on clinical E. faecalis isolates in Shiraz, Iran, showed that the prevalence rates of the asa1, esp, gelE, cylA, and hyl genes were respectively 100%, 94.1%, 80.4%, 64.7%, and 51% (47), respectively. The reported rates are higher than the rates obtained in our study. Moreover, the prevalence of the asa1 gene was higher in the present study than in some previous studies (48-50).
The high prevalence of the asa1 gene in the present study can facilitate the exchange of virulence and antimicrobial resistance genes in our setting. The second most prevalent genes in the present study were the asa1 and esp genes, with a prevalence rate of 74%. The high prevalence of the esp gene in our study is in line with the findings of some other studies (51-53). Moreover, our findings showed that the E. faecalis isolates carried more virulence genes compared to the prevalence reported in some previous studies (18, 28, 48, 54, 55). This difference may be due to the differences in the participants’ characteristics and geographical dispersion. Considering the remarkable role of virulence factors in the adhesion, colonization, and biofilm formation ability of bacteria, the high prevalence of Enterococcus species which carry virulence factors can be associated with more severe infections (47).
The genetic diversity analysis revealed 25 ERIC types, and the dendrogram analysis showed that most E. faecalis isolates (n = 35) were clustered into ten clusters (ie, clusters A-J). These clusters were further clustered into two major clusters H with eight isolates and J with ten isolates, as well as eight minor clusters, each with 2 - 3 isolates. These findings can imply epidemiological relations among some circulating MDR E. faecalis isolates in the study setting. Moreover, the genetic diversity analysis showed great genetic diversity among the isolates not included in the two major clusters. Great genetic diversity among the isolates can improve the survival of different E. faecalis strains in the study setting. Several studies in Iran and other countries have also reported the genetic diversity of the E. faecalis isolates. For example, a study in Isfahan, Iran, found fourteen ERIC types among 53 E. faecalis isolates (45). Another study in Indonesia showed that all E. faecalis isolates had > 60% similarity and reported five clusters (19). Moreover, a study on patients with burn injuries in Tehran, Iran, found 34 ERIC types among 57 Enterococcus isolates (18). These contradictory findings can be attributed to the high heterogeneity of E. faecalis due to differences in its nucleotide sequence.
The antimicrobial resistance analysis in the two major clusters H and J showed the greatest antimicrobial resistance to antibiotics E, RI, and TET. However, there was no significant difference between these two clusters in terms of the phenotypic and the genotypic patterns of antimicrobial resistance. However, the prevalence of the asa1 virulence gene was significantly higher in cluster J than in cluster H. Different bacterial colonies may have different virulence genes and factors; hence, they can pose more problems in managing their associated infections. Given the critical role of virulence factors in the virulence of E. faecalis, the high prevalence of these strains can complicate patients' conditions. One of the limitations of this study was the small sample size. Moreover, the study was conducted in one hospital, which resulted in not having enough bacterial isolates in all clusters. Further studies are recommended to address larger sample sizes and several hospitals.
5.1. Conclusions
This study showed the high prevalence of MDR and antibiotic resistance to antibiotics E, RI, and TET among the E. faecalis isolates obtained from the stool samples of patients in a hospital in the center of Iran. Most E. faecalis isolates were susceptible to LZD. The most prevalent antimicrobial resistance genes were ermB, aph(2′′)Ia, and aac(6′)-Ie, while the most prevalent virulence genes were gelE, asa1, and esp. The ERIC-PCR analysis also revealed that the isolates were relatively heterogeneous as such they were clustered into two major and eight minor clusters with no significant difference between the two major clusters in terms of the phenotypic and genotypic patterns of antimicrobial resistance and virulence genes.