1. Background
Streptococcus agalactiae or group B Streptococcus (GBS) is serologically distinguished from other species in the genus Streptococcus. It is an encapsulated, facultative anaerobic, nutritionally fastidious diplococcus, with ß-hemolytic activity on blood agar plates. The unique characteristics of GBS include hydrolysis of sodium hippurate, resistance to bacitracin, positive results on the CAMP co-hemolytic test. In pregnant women, GBS causes bacteriuria and chorioamnionitis. It can also cause pneumonia, meningitis, endocarditis, sepsis, and bacteremia in infants. Infections sometimes begin inside the uterus (1, 2). Adhesion, as the first stage of GBS colonization, is mediated by adhesion factors present on the bacterial surface, allowing for the attachment of GBS to extracellular matrix proteins and epithelial cells of the intestinal and genital tracts. Moreover, adhesion factors can increase invasion either through destruction of the epithelial cell layer or disarrangement of the epithelial cytoskeleton, which facilitates paracellular transport.
Fibrinogen-binding proteins (FbsA and FbsB) and laminin-binding protein (Lmb) are major factors in adherence to the extracellular matrix. FbsA mediates the adhesion of GBS to host cells, while FbsB promotes the invasion of GBS. The Lmb adhesion appears to play a prominent role in bacterial attachment to the extracellular matrix by facilitating GBS translocation across the intestinal epithelium and the blood-brain barrier (3). A new, recently discovered GBS fibronectin-binding protein, called streptococcal fibronectin-binding protein A (SfbA), has been shown to be highly conserved in GBS, contributing to cellular invasion rather than adherence (4). The SfbA adhesion has been reported to be directly involved in fibronectin binding and human brain microvascular endothelial cell (HBMEC) invasion. Another study exhibited the significant role of SfbA in the invasion of astrocytes, which have a physical connection with the brain endothelial cells (5). Moreover, SfbA contributes to GBS invasion of vaginal and cervical epithelial cells, possibly promoting GBS colonization and niche settlement in the vagina (4).
Considering the GBS pathogenicity and the occurrence of antibiotic resistance, various strains of this bacterium are emerging, whose frequencies differ across geographical areas. Therefore, it is important to investigate genomic polymorphisms and determine the primary colonization genes of GBS isolates in various regions to prevent the occurrence of drug resistance and high-risk infections in pregnant women and infants. Specific molecular techniques can be effective in identifying the species diversity in epidemiological studies.
2. Objectives
Regarding the scarcity of national and regional data, the current study aimed to determine the genetic relatedness of GBS isolates collected from pregnant women using the BOX-PCR fingerprint technique and to determine the presence of three central colonization-related genes (fbsA, fbsB, and lmb), as well as a novel gene (sfbA), using a multiplex PCR assay.
3. Methods
3.1. Study Design
In this descriptive cross-sectional study (January 2021- July 2021), a total of sixty pregnant women with urinary tract infections (UTIs) admitted to a hospital in Tehran, Iran, were included. The infection of all participants had been confirmed by a specialist based on demonstrating a colony count of more than 105 CFU/mL in their urine sample. After describing and clarifying the project, oral consent obtained from participants.
3.2. Bacterial Isolates and Materials
Sixty bacterial strains from urine (16 isolates) and placenta (44 isolates) samples of target participants were isolated at the microbiology laboratory of the hospital and confirmed as GBS. Then GBS isolates were further examined by gram staining, colony morphology, ß-hemolytic activity, and conventional biochemical tests such as, sodium hippurate hydrolysis, CAMP test, and resistance to bacitracin. Furthermore, GBS isolates were molecularly confirmed by amplifying the dlts gene (6). Streptococcus agalactiae ATCC 12403 and Streptococcus pyogenes ATCC 1244 were applied as positive and negative controls, respectively. All GBS isolates were grown on a 5% sheep Blood agar at 37°C under the micro-aerobic atmosphere and then inoculated in BHI broth (Brain Heart Infusion broth) containing 25% glycerol and transferred to -20°C until the time of molecular examinations. All chemical materials and culture media were prepared from Merck (Darmstadt, Germany).
3.3. Genomic DNA Extraction
Genomic DNA was extracted via a commercial Genomic DNA extraction Kit (cat No: DM05050, Gene Transfer Pioneer, Pishgaman Co, Iran) based on the manufacturer’s procedure.
3.4. Molecular Detection of Colonization-Associated Genes
The colonization associated genes; ftsA, ftsB, lmb, and sfbA were simultaneously investigated using the multiplex PCR technique. The primers used for current search were extracted from related articles (4, 7) (Table 1) and their validity was checked using BlastN algorithm, available at NCBI. The primers were synthesized by Macrogen Inc; South Korea. A positive control, confirmed by sequencing, was considered for all target colonization genes. The multiplex PCR was performed with two μL of DNA (50 ng) in a final volume of 20 μL containing ten μL of 2X master mix with standard buffer and one μL of each four primer pairs. The cycling program was as follow: Primary denaturation at 94°C for 3 min (initial denaturation), followed by 30 cycles (30 s at 94°C, 30 s at 58°C, 60 s at 72°C) and a final extension at 72°C for 5 min, in a thermocycler (Bio-rad T100, Inc). The obtained multiplex PCR products were separated via 1.5% agarose gel (m/v) and an electrophoretic cell (at 110 V for 60 min) and subsequently, the bands were visualized by the ultraviolet (UV) transilluminator (PoteinSimple, Red imager SA-1000).
| Gene Category | Function/Annotation | Primer Sequence (5’-3’)a | Size of Amplicon (bp) |
|---|---|---|---|
| fbsA | Fibrinogen-binding protein | F:TGTAGCTAATGGACCGATGTT | 156 |
| R:TTTTCATTGCGTCTCAAACC | |||
| fbsB | Fibrinogen-binding protein | F:ACAACTGCGGAAATGACCTC | 186 |
| R:ACGAGCGACGTTGAATTCTT | |||
| lmb | Laminin-binding protein | F:AGTCAGCAAACCCCAAACAG | 397 |
| R:GCTTCCTCACCAGCTAAAACG | |||
| sfbA | Fibronectin-binding protein | F: CTACTCCTATCTGCTCACCCTGTA | 288 |
| R: GTTGAACCAGGAAGGATAGTACGG |
Primers Used to Amplify Colonization-Associated Genes in Group B Streptococcus Strains
3.5. BOX PCR Fingerprinting
To determine the genetic polymorphism of isolates, BOX-PCR fingerprinting using the primer BOXA1R (5′-CTACGGCAAGGCGACGCTGACG-3′) was applied (8). The 20 μL reaction mixture included ten μL of 2X master mix (Ampliqon manufacture, Danish) (containing; buffer, deoxy nucleoside triphosphate mix, and Taq DNA polymerase, and Mg+2), one μL of primer, and two μL of DNA template (50 ng). PCR condition comprised initial denaturation at 95°C for 3 min, followed by 60 cycles of denaturation at 94°C for 30 s, primer annealing at 45°C for 30 s, and extension at 72°C for 80 s, with a final extension at 72°C for 5 min. The PCR products were separated by electrophoresis using 1.5% agarose gel in a 1x TBE buffer. The ethidium bromide stain and a UV transilluminator were applied to visualize PCR products. A 100bp DNA ladder (Sinacolon Corporation, Iran) was considered as molecular weight marker.
3.6. Cluster Analysis
The banding patterns produced by BOX-PCR were analyzed via NTsys-pc version 2.02. Cluster analysis was done through an unweighted pair group method (UPGMA) and Jaccard's similarity coefficient (8). The dendrogram was cut in the similarity coefficient of 0.6, and the GBS isolates were classified.
4. Results
The demographic information of participants is shown in Table 2. The age of women was ranging from 19 to 46 years. Of the 60 women included in the study, 13 were first deliveries, 5 were second deliveries, and 42 were third deliveries. The gestational age of women was as follows; 43 women in the first trimester, 10 in the second trimester and 3 in the third trimester, three other in the fourth trimester, and 1 in the fifth trimester. Out of 60 women studied, only three who had diabetes mellitus type 2 were taking metformin and glibenclamide (hypoglycemic drugs). None of them had received antibiotics two weeks before study, and also, they did not experience previous spontaneous abortion.
| Patient No. | Age | Month of Abortion | Number of Delivery | Source of Isolation (Urine/Placenta) | BOX PCR Genotype | Patient No. | Age | Month of Abortion | Number of Pregnancy | Source of Isolation (Urine/Placenta) | BOX PCR Genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 35 | 2 | 1 | U a | 6 | 31 | 33 | 1 | 2 | U | 2 |
| 2 | 33 | 1 | 1 | P b | 6 | 32 | 39 | 1 | 3 | U | 2 |
| 3 | 27 | 1 | 1 | P | 6 | 33 | 38 | 1 | 3 | P | 2 |
| 4 | 29 | 1 | 1 | P | 6 | 34 | 38 | 1 | 3 | P | 2 |
| 5 | 19 | 1 | 1 | P | 6 | 35 | 41 | 2 | 3 | P | 2 |
| 6 | 19 | 1 | 3 | P | 6 | 36 | 40 | 2 | 1 | P | 2 |
| 7 | 21 | 1 | 3 | P | 6 | 37 | 36 | 1 | 3 | P | 4 |
| 8 | 26 | 4 | 3 | P | 6 | 38 | 33 | 1 | 3 | P | 4 |
| 9 | 27 | 3 | 3 | P | 6 | 39 | 34 | 1 | 3 | U | 6 |
| 10 | 42 | 2 | 2 | P | 6 | 40 | 32 | 1 | 3 | U | 6 |
| 11 | 41 | 1 | 3 | U | 6 | 41 | 29 | 1 | 2 | P | 2 |
| 12 | 39 | 1 | 3 | P | 6 | 42 | 28 | 1 | 3 | P | 1 |
| 13 | 36 | 1 | 3 | P | 4 | 43 | 26 | 1 | 3 | P | 2 |
| 14 | 36 | 2 | 3 | P | 5 | 44 | 29 | 1 | 3 | U | 6 |
| 15 | 36 | 2 | 3 | P | 5 | 45 | 22 | 1 | 1 | P | 5 |
| 16 | 35 | 1 | 3 | P | 4 | 46 | 21 | 1 | 3 | U | 5 |
| 17 | 33 | 1 | 3 | P | 4 | 47 | 19 | 1 | 3 | P | 1 |
| 18 | 22 | 3 | 3 | P | 4 | 48 | 26 | 3 | 3 | U | 4 |
| 19 | 21 | 1 | 1 | P | 3 | 49 | 27 | 2 | 3 | P | 1 |
| 20 | 27 | 4 | 1 | P | 4 | 50 | 45 | 4 | 3 | U | 4 |
| 21 | 29 | 5 | 3 | U | 4 | 51 | 40 | 1 | 3 | U | 2 |
| 22 | 44 | 2 | 1 | P | 3 | 52 | 36 | 1 | 2 | P | 2 |
| 23 | 43 | 2 | 1 | P | 3 | 53 | 33 | 1 | 3 | P | 2 |
| 24 | 46 | 2 | 1 | P | 3 | 54 | 31 | 1 | 3 | P | 2 |
| 25 | 19 | 1 | 3 | P | 2 | 55 | 30 | 1 | 2 | P | 2 |
| 26 | 20 | 1 | 3 | P | 2 | 56 | 29 | 1 | 1 | P | 2 |
| 27 | 22 | 1 | 3 | P | 2 | 57 | 28 | 1 | 3 | P | 2 |
| 28 | 24 | 1 | 3 | U | 1 | 58 | 32 | 1 | 3 | U | 2 |
| 29 | 26 | 1 | 3 | U | 2 | 59 | 37 | 1 | 3 | P | 2 |
| 30 | 29 | 1 | 3 | U | 2 | 60 | 39 | 1 | 3 | P | 2 |
Demographic Data of Pregnant Women Admitted to the Hospital and Compilation of BOX- PCR Data for Clinical Group B Streptococcus Isolates
4.1. Distributional Characteristics of Colonization-Associated Genes
The BLAST search for sequences of each understudied colonization gene against sequences in the nucleotide database revealed that they belong to genus group B Streptococcus and showed maximum identity with S. agalactiae strain FDAARGOS 670. Based on results obtained from multiplex PCR and related primers, all isolates contained four target colonization genes, fbsA, fbsB, lmb, and sfbA, simultaneously.
4.2. BOX Fingerprinting of Clinical GBS Isolates
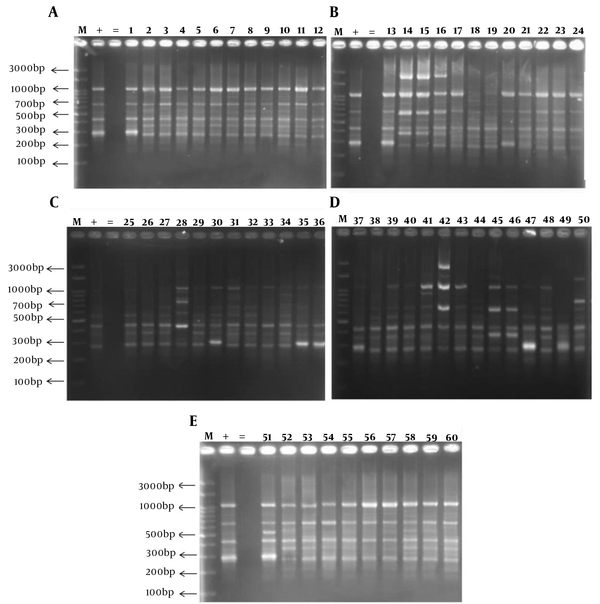
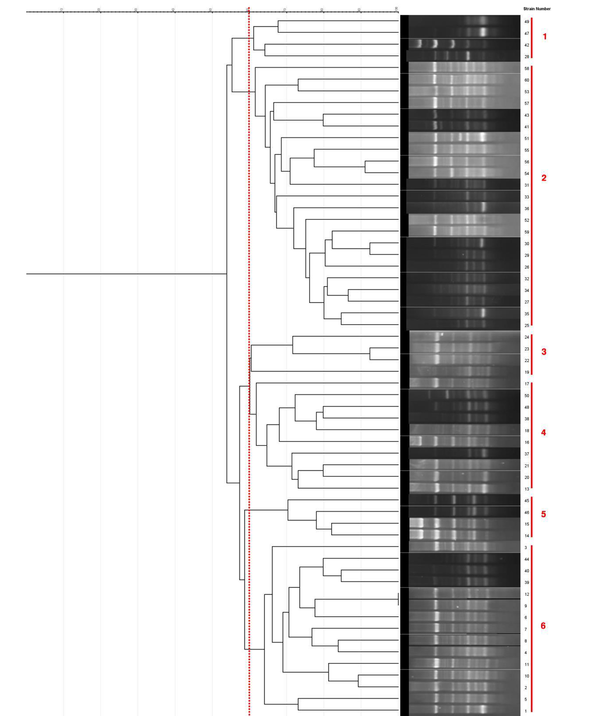
BOX-PCR fingerprinting using the BOXA1R primer produced 4 to 11 bands ranging from 300 - 3000 bp (Figure 1A- E). Most of the isolates were in common in four bands per pattern. The most specific PCR products of understudied GBS isolates were as follows: 300, 400, 700, and 1000 bp. According to the dendrogram, all clinical GBS isolates were divided into 6 clusters at 60% level as follow; three dominant genotypes including 10 to 23 isolates and three other distinctive patterns. (Figure 2)
Dendrogram obtained by comparing BOX-PCR fingerprinting patterns (cutoff 60%) of 60 clinical group B Streptococcus isolates from pregnant women (UPGMA analysis, Jaccard coefficient). 60 group B Streptococcus isolates containing all studied colonization genes (fbsA, fbsB, lmb, and sfbA) were distinguished into 6 groups by using BOXA1R primer.
5. Discussion
GBS is known as a part of the intestinal and vaginal microbiota in 15 - 30% of healthy women; nonetheless, it has an extreme capacity to cause invasive infections. This bacterium is a significant cause of infections in infants, pregnant women, and adults with underlying diseases. This pathobiont, which grows in healthy individuals, has maintained its potential virulence during coevolution with humans. It is well established that this bacterium is one of the most life-threatening pathogens in neonates and the elderly. Moreover, it has been reported that one in every 10 newborn infants acquires GBS vertically while passing through the birth canal or soon afterward (1, 2, 9, 10). In 1996, the Centers for Disease Control and Prevention (CDC) issued guidelines for the prevention of GBS before delivery, recommending screening for GBS colonization in prenatal women. Although previous studies reported that up to 50% of infants with GBS are born to carriers without clinical risk factors, the CDC guidelines were revised in 2002, and bacteriological screening became mandatory for all pregnant women at 35 - 37 weeks of gestation. Therefore, for improving public health, it is important to quickly identify and differentiate GBS strains (11).
The GBS adherence and colonization, as complex multifactorial functions, guarantee the maintenance of this pathobiont in human ecosystems. Over the last two decades, molecular adherence mechanisms have been further studied in GBS, leading to a significant increase in our understanding of disease progression. The GBS adhesion factors may present new targets for the emergence of novel treatments and preventive actions to control and prevent invasive GBS infections in newborns and adult patients, as the frequency of vaginal-rectal colonization of GBS changes significantly across geographical regions, even in a single country. Data regarding the source of infection and relatedness of isolates can be beneficial for preventive strategies and selection of the best treatment (12). In the current study, besides assessing the concomitant presence of central colonization genes through multiplex PCR assay, the amenability of BOX-PCR fingerprint technique was investigated to differentiate GBS isolates collected from urinary and placental samples of pregnant women, referred to one of the hospitals of Tehran, Iran.
Significant adhesions that mediate the colonization of GBS on host cells, including fibrinogen-binding proteins (Fbs A and B), laminin-binding protein (Lmb), and a recently identified fibronectin-binding protein (SfbA), were concomitantly studied in the GBS isolates using the multiplex PCR assay. The results revealed that all GBS isolates harbored the studied colonization genes. Therefore, colonization-related genes showed a similar distribution among isolates originating from two different types of clinical samples (urine and placenta). The positive rates of colonization genes, including fbsA and fbsB, were identical to those reported in previous studies, except for lmb; this result is consistent with the finding that most human isolates contain fbsA and fbsB genes (7, 13, 14). Additionally, previous studies have indicated the concomitant harboring of three or more than three colonization-related genes in the GBS isolates (15, 16). However, the distribution rate of genes for surface-localized proteins, such as fbsA and fbsB, was different, while lmb was found in all the isolates. Discrepancies in the distribution and abundance of colonization-related genes are probably related to regional differences; therefore, identification and survey of dominant colonization-associated genes in different geographical regions are suggested as an appropriate tool to design efficient vaccines for the prevention of GBS infections in pregnant women (17).
The sfbA gene was also harbored by all isolates in the current study. Similar studies have earlier reported that sfbA gene is present in GBS and highly conserved (4, 18). The present study is the first report of sfbA gene identification among GBS isolates collected from pregnant women in Iran. The BOX primer sequence was used in a PCR assay to determine differences in the number and distribution of this bacterial repeat sequence in the genomes of clinical GBS isolates. Although among different molecular techniques for typing bacterial strains, pulse-field gel electrophoresis (PFGE) is known as the superior technique for bacteria, it is both labor-intensive and costly (19). In contrast to PFGE, BOX-PCR is known as the most commonly applied method in biogeographic studies of bacterial isolates. Researchers have demonstrated that this technique is rapid, vastly discriminative, and reproducible (8). Therefore, BOX-PCR was identified as a selective typing method for GBS isolates in this study.
According to the current findings, the most frequent genotypes were genotypes 2, 4, and 6 found in 23 (38.33%), 10 (16.66%), and 15 (25%) isolates, respectively. The subsequent genotypes, including genotypes 1, 3, and 5, were each found in 4 (6.66%) isolates. The isolation source in most GBS isolates (73.33%) was the placenta of patients, while the remaining isolates (26.66%) were obtained from the urinary samples. Except for genotype 3, other genotypes were found in isolates from both samples; this finding revealed that isolates from the placental and urinary samples were highly heterogeneous.
To the best of our knowledge, there are no publications in Iran or other countries on the molecular typing of GBS using BOX-PCR. Nevertheless, genotyping of GBS using other fingerprint techniques has been reported (20, 21). Also, the effectiveness of BOX-PCR in determining genetic relatedness has been investigated for other bacterial species. For instance, the eligibility and reproducibility of BOX-PCR and ERIC-PCR were used as molecular typing tools for the genetic discrimination of Enterococcus faecalis.
Besides, other researchers have reported the potential of BOX-PCR to differentiate Proteus mirabilis strains from clinical Pseudomonas aeruginosa isolates (22-25). In other studies from other countries, BOX-PCR and ERIC-PCR were introduced as powerful surveillance techniques for studying the genetic relatedness of Leptospira, Fusarium oxysporum, and Salmonella enterica isolates from patients (8, 26, 27). Samples included in this research were collected from a hospital in Tehran, Iran during the six-month period from January to July 2021. Thus, the results of this study may not be generalized to other areas. More investigations are required on samples collected from different regions of Iran and during various seasons.
5.1. Conclusions
Based on the results of the present study, four important colonization-related genes, including fbsA, fbsB, lmb, and sfbA, were concomitantly detected in all 60 GBS isolates. The BOX-PCR fingerprint technique could discriminate GBS isolates, collected from placental and urinary samples of pregnant women, into six different groups. Since the analysis of phenotypic features does not present a classification system, this DNA fingerprint technique can be a rapid and useful tool for identifying GBS isolates.