1. Background
Candidiasis is known as an opportunistic fungal infection and is the third cause of healthcare-related infections with high morbidity in immunocompromised patients (1). Candidiasis, as a local yeast infection, is typically caused by Candida albicans (2). Candida species are frequently detected in patients with mucosal, skin, and nail infections (3-5). The invasive and systemic diseases caused by Candida species in susceptible host populations have a high mortality rate, despite medical and surgical treatment (6). Candidiasis in the vagina is commonly called vaginal candidiasis or candidal vaginitis, caused by Candida species at least in 20 - 25% of cases (7).
It is a serious health problem to enhance the resistance of C. albicans to antifungal agents, mainly to azoles. Approximately 5% of women at least four times a year are infected due to the resistance of candida species to antifungal agents (8). Therefore, there are serious challenges to drug resistance in the treatment of this disease (9). Accordingly, it is essential to identify new antifungal agents against resistant species to effectively treat patients affected by these species. Different molecular mechanisms have been identified in drug resistance to fluconazole in C. albicans (10). One of the important mechanisms causing the formation of azole resistance is the reduction of intracellular drug accumulation, which is associated with the expression and enhancement of the genes involved in the efflux pump system, such as Candida drug resistance 1 (CDR1) and Candida drug resistance 2 (CDR2) genes (11). The ERG11 gene encodes lanosterol 14α-demethylase, which is a target of fluconazole (12, 13).
Currently, nanoparticles (NPs) are commercialized as antimicrobial and antifungal agents (14, 15). Zinc oxide nanoparticles (ZnO-NPs) function through membrane integrity disruption by the production of reactive oxygen species and are suggested as a candidate for the elimination of C. albicans biofilm (12). Recently, it has been shown that the formation of biofilms of C. albicans infections is necessary for binding to vaginal cells, which is known as the first step in the development of infection (13). The important genes are involved in adhering C. albicans to mucosal surfaces and belong to the agglutinin-like sequence (ALS) and hyphal wall protein (HWP) family (16).
2. Objectives
The present study aimed to investigate the changes in the expression levels of ERG11, CDR2, HWP1, and ALS1 genes in C. albicans isolates after the investigation of the optimal concentrations of ZnO-NPs and fluconazole by real-time polymerase chain reaction (PCR).
3. Methods
3.1. Specimens and Cultivation and Identification of Yeasts
In this case-control study, the population included 120 suspected women with vaginitis randomly selected from patients that referred to the health centers in Qom province, Iran, within February 2019 to May 2020. Sabouraud dextrose agar (Biolife, Italy) containing chloramphenicol was used to incubate vaginal specimens collected using sterile moisture swabs. After 24 to 48 hours of incubation at 30°C, the isolates were detected by both the morphological method on CHROM agar Candida (CHROMagar Company, France) and PCR amplification of the internal transcribed spacer (ITS) region with 5´-TCCGTAGGTGAACCTGCG-3´ as a forward primer and 5´-TCCTCCGCTTATTGATATGC-3´ as a reverse primer, which produces a 535 bp band on agarose gel (17). For the extraction of the genomic deoxyribonucleic acid from the isolates, the SinaClon DNPTM kit (SinaClon, Iran) was utilized as directed by the manufacturer’s protocol. The PCR was used to amplify the ERG11, ALS1, HWP1, and CDR2.
3.2. Preparation of ZnO-NPs
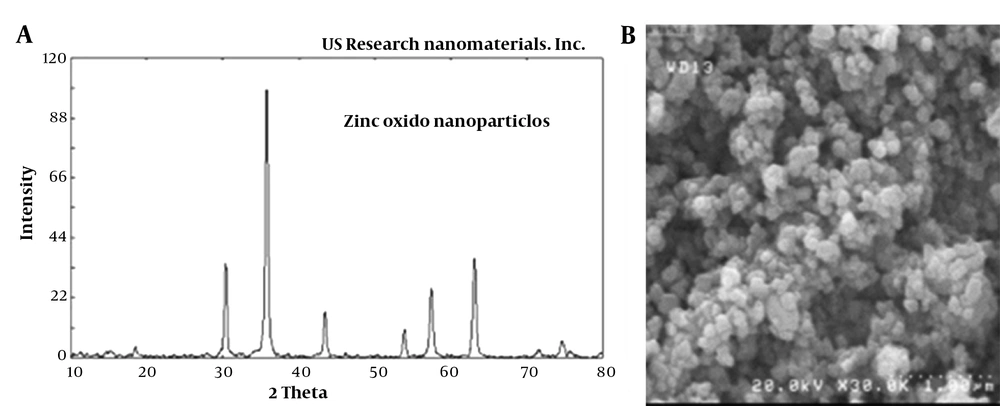
A wet chemical method was performed to synthesize the ZnO-NPs. Briefly, the aqueous sodium hydroxide solution (0.8 mol/L) was gradually added to the 10 mL of zinc chloride solution (0.4 mol/L) until proper dissolution. After 2 hours of room temperature incubation, the procedure was followed by five-time washing steps with distilled water. For obtaining ZnO-NPs, the final precipitates were dried at 400°C (18). The structural and morphological characterization of ZnO-NPs was examined by X-ray diffraction (XRD; Shimadzu, Kyoto, Japan) and scanning electron microscope (SEM; Zeiss DSM 960A, Carl Zeiss, Oberkochen, Germany) at the Central Laboratory, University of Tehran. The average particle size of the synthesized NPs was 35 nm, and their shape and porosity (%) were almost uniform (spherical and white) and > 99, respectively (Figure 1). According to the topographical perspective, NPs are more or less spherical in nature, clustered together, and the surface of the aggregates appears to be rough. The shape of NPs has a significant impact on pathogens because spherical NPs tend to be very potent during antibacterial activity owing to their ability to easily penetrate into the cell wall of pathogens (19).
3.3. Determination of Candida albicans Isolates Sensitivity to ZnO-NPs and Fluconazole
Based on the recommendations of the Clinical Laboratory Standards Institute (CLSI document M44-A2), the resistance to fluconazole was determined by the disk diffusion assay. Accordingly, to carry out the test, massive culture was performed by sterile swabs impregnated with fungal suspension in accordance with an opacity of half the McFarland tube on a Mueller-Hinton agar medium. After drying the plate at 37°C for 15 minutes, a fluconazole disk was applied. The plates were incubated for 24 hours at 30°C, and the diameter of the inhibition zone was measured. The isolates with zone diameters of ≤ 14, 15 - 18, and ≥ 19 mm were reported as resistant, semi-sensitive, and sensitive, respectively. Moreover, the minimum inhibitory concentration (MIC) of the ZnO-NPs was evaluated by the broth microdilution method according to the CLSI recommendations described in a previous study (20).
Accordingly, 100 μL of Sabouraud dextrose broth medium was added to each plate well, and 100 μL of fluconazole (1024 μg/mL) and nano-ZnO (1000 μg/mL) were both separately and simultaneously added to the first well. Then, 100 μL of the first well was taken and poured into the second, and a procedure was carried out successively. Afterward, 10 μL of fungal suspension was added to each well. Two wells were used as positive and negative controls. A medium containing 10 μL of fungal suspension served as a positive control; however, a medium containing an antifungal with no growth served as a negative control. The fungal growth was measured using a spectrophotometer at 600 nm after a 24-hour incubation period. The MIC was defined as the lowest dose of an antifungal compound that inhibited the growth of the tested fungi (20).
3.4. Analysis of ERG11, ALS1, HWP1, and CDR2 Expression Levels in Candida albicans Isolates
For the evaluation of CDR-2, ERG11, ALS1, and HWP1 expression, after MIC analysis, the C. albicans isolates were given sub-MIC doses of ZnO-NPs, fluconazole, and a combination of ZnO-NPs and fluconazole. Ribonucleic acid (RNA) was extracted from the fluconazole-resistant C. albicans isolates by the phenol-chloroform extraction method, as previously mentioned (21, 22). Briefly, the harvested cells with an OD 600 of approximately 0.6 were centrifuged at 6000 rpm for 5 minutes, resuspended in 1 mL of triazole buffer, and combined by vortexing. Then, the suspension was vigorously mixed for 5 minutes upon the addition of acid-washed glass beads and 1 mL of phenol: chloroform (1: 1). After centrifugation for 15 minutes at 1000 rpm, the top aqueous phase was transferred to a fresh tube.
After two rounds of ethanol wash, the RNA was in 100 µL diethylpyrocarbonate (DEPC) water (Invitrogen, Thermo Fisher Scientific, USA) and stored at -70ºC until required. The complementary deoxyribonucleic acid (cDNA) synthesis was conducted by BioFactTM RT-Kit (BioFACT, Daejeon, Korea) following deoxyribonuclease enzyme treatment, in accordance with the manufacturer’s instruction. Reverse transcription PCR was performed in a 20 μL reaction containing 5 μL of template cDNA, 0.5 μL of each specific primer for the ERG11, ALS1, HWP1, and CDR2 genes, and 4 μL of DEPC water. The initial denaturation step was performed at 95°C for 10 minutes, followed by 40 cycles of denaturation at 93°C for 10 seconds, the annealing step at 50°C for 40 seconds, and extension at 72°C for 30 seconds, followed by a final extension step at 72°C for 3 minutes. The internal control was the ACT1 gene. The primers designed by the Primer3 web-based software (version 0.4.0) (http://frodo.wi.mit.edu/primer3) were employed for real-time PCR analysis. The specificity of the primers was confirmed using the BLAST search on the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast.cgi), which is shown in Table 1.
| Primers | Sequence (5’ - 3’) | Tm (°C) | Size Band | Accession Number |
|---|---|---|---|---|
| CDR2-F | GCTAGACGAAAACCCATGG | 50 | 213 bp | XM_718076.2 |
| CDR2-R | ATGTTGCCGTTGAATGGAC | 50 | ||
| ERG11-F | CAAAAATTACCATCAGTCAATAACAC | 50 | 280 bp | XM_711668 |
| ERG11-R | CAAACCCATAATCAACTTCATCAG | 50 | ||
| HWP1-F | CTGCTCAACTTATTGCTATCGC | 51 | 152 bp | XM_704869.2 |
| HWP1-R | TTGTTGTTGTGGGTAATCATCA | 51 | ||
| ALS1-F | TGCCATATCATACTACCACAACTG | 51 | 129 bp | XM_712984.2 |
| ALS1-R | CAGTTGGATTTGGCAGTGG | 51 | ||
| ACT1-F | TGGTATGGGTCAAAAAGATTC | 50 | 172 bp | XM_019475182.1 |
| ACT1-R | TGGATGTTCTTCTGGAGCA | 50 |
3.5. Statistical Analysis
The SPSS software (version 22; Chicago, IL, USA) was employed for all statistical analyses. The student’s t-test was used to compare and assess the control and treatment groups in various tests. The data with p-values less than 0.05 were considered statistically significant. Graphs were depicted by GraphPad Prism software (version 8).
4. Results
4.1. Susceptibility Testing of Candida albicans Isolates
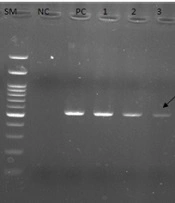
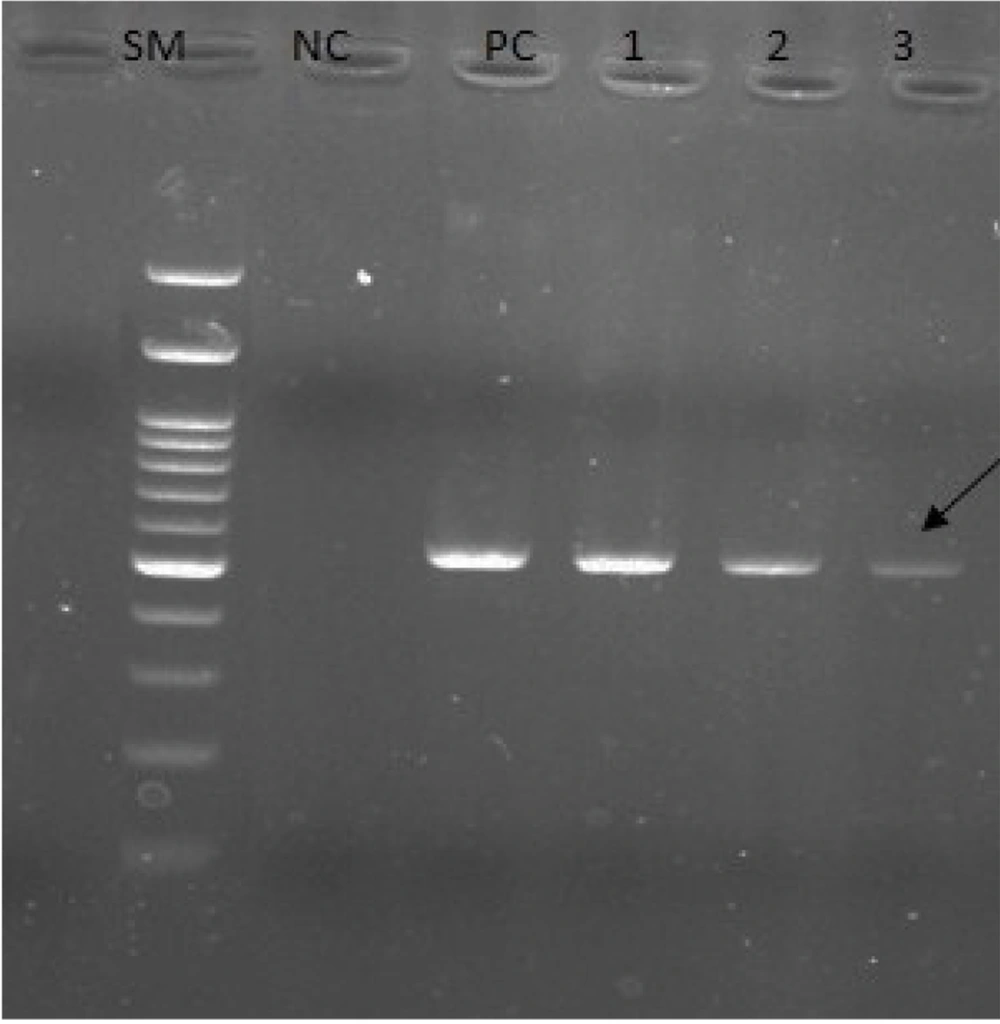
In a previous study, 41.6% of C. albicans species (50/120) were isolated from 120 patients suspected of candida infection with or without vulvovaginal symptoms. The 50 identified isolates were verified by performing PCR using specific primers for the ITS region (Figure 2). Out of 50 isolates, 13 (26%) isolates were resistant to fluconazole with an inhibition zone broader than ≤ 14 mm in diameter; however, 37 (76%) isolates were fluconazole sensitive (inhibition zone > 14 mm). The XRD and SEM were used to examine the crystalline structure and detect the ZnO-NPs (Figure 1). According to the obtained findings, the average particle size of the synthesized NPs was 35 nm, and the shape and porosity of the NPs (%) were almost uniform (spherical and white).
Electrophoresis of polymerase chain reaction products of Candida albicans isolates using internal transcribed spacer (ITS)-specific primers (SM: 100 bp deoxyribonucleic acid size marker; NC, negative control; PC, positive control; lanes 1 - 3, ITS-positive isolates). PCR amplification using these primers produces a 535 bp product.
4.2. Determination of MIC of ZnO-NPs and Fluconazole Against Candida albicans Isolates
The MIC values of ZnO-NPs and fluconazole on C. albicans isolates were 19.9 and 7 µg/mL, respectively (Table 2). The comparison of results from the combination of ZnO-NPs with fluconazole showed the almost satisfactory effects of this compound. After examining its pharmacodynamic and pharmacokinetic effects, it is possible to comment with more certainty on the antifungal effects of this combination.
| Strains (No. of Isolates) | Antifungal Agent | MIC Range (µg/mL) | MIC50 (µg/mL) | MIC90 (µg/mL) |
|---|---|---|---|---|
| Candida albicans (n = 50) | Fluconazole | 0.031 - 128 | 7 | 14.1 |
| ZnO-NPs | 0.02 - 296 | 19.9 | 44.7 |
Abbreviations: MIC50, minimum inhibitory concentration 50%; MIC90, minimum inhibitory concentration 90%; ZnO-NPs, zinc oxide nanoparticles.
4.3. Real-Time PCR Analysis
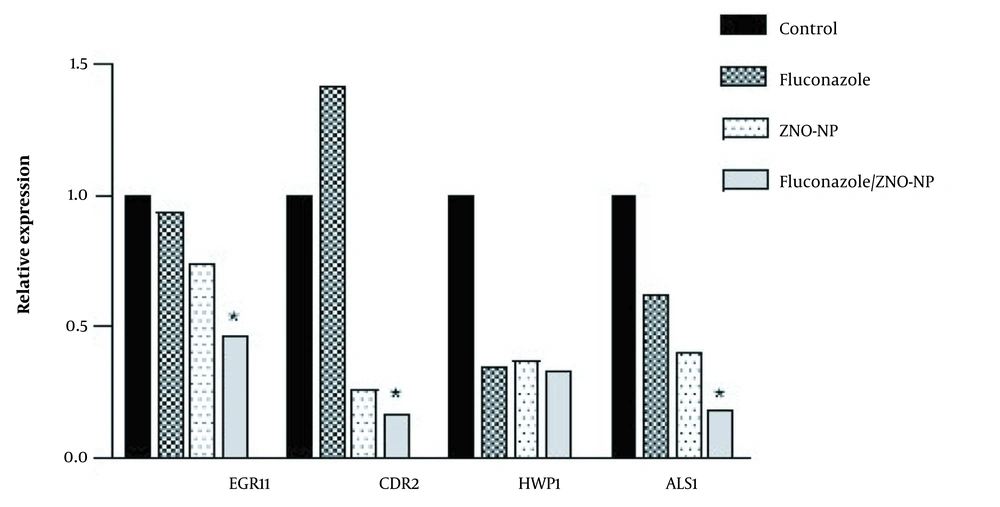
In this study, the changes in the expression levels of ERG11, CDR2, HWP1, and ALS1 genes in the fluconazole-resistant C. albicans isolates were evaluated using ACT1 as the reference gene (Figure 3). The obtained results showed the expression of HWP1 and ALS1 genes decreased by 2.84 and 1.62 times after taking sub-MIC values of fluconazole (P < 0.05), respectively. The expression analysis showed that, after treatment with fluconazole, the expression of CDR2 increased by 1.42-fold; nonetheless, the expression of the ERG11 gene did not differ significantly. Moreover, the statistical analysis of expression data showed that the gene expression of ERG11, CDR2, HWP1, and ALS1 was significantly downregulated in C. albicans isolates after treatment with ZnO-NPs by 1.35, 3.8, 2.7, and 2.49 times, respectively.
The expression data revealed that the treatment of isolates with the combination of ZnO-NPs and fluconazole downregulated the expression levels of ERG11, CDR2, HWP1, and ALS1 in comparison to those of the control group by 2.1, 5.9, 3, and 5.5 times, respectively (Figure 3). The aforementioned data showed that in comparison to the fluconazole group, the combination of ZnO-NPs and fluconazole caused a significant decrease in the expression levels of ERG11, CDR2, and ALS1 (P < 0.05). Although the expression level of HWP1 was downregulated in all three treatment groups, there was no significant difference between them (P > 0.05).
Relative expression of EFG11, CDR2, HWP1, and ALS1 in Candida albicans; isolates treated with fluconazole, zinc oxide nanoparticles (ZnO-NPs), and their combination; total ribonucleic acid extracted and reverse transcribed to complementary deoxyribonucleic acid for further real-time quantitative polymerase chain reaction to detect gene expression levels (∗ P < 0.05 compared to the fluconazole alone group).
5. Discussion
This study indicated evidence of the antifungal activity of ZnO-NPs with fluconazole on C. albicans isolates and their effects on the expression levels of ERG11, CDR2, HWP1, and ALS1 genes. Different sub-MIC values of ZnO-NPs, fluconazole, and a mixture of ZnO-NPs and fluconazole were used to treat fluconazole-resistant Candida isolates. In the present study, the expression of ERG11, CDR2, HWP1, and ALS1 in isolates treated with combining ZnO-NPs and fluconazole was downregulated in comparison to that of the control group. The present study indicated that, when compared to using ZnO-NPs or fluconazole alone, the synergistic activity of ZnO-NPs disc with fluconazole had a significant effect on isolated C. albicans. The aforementioned results are in line with the results of previous studies (17, 20).
Several studies revealed that the expression of the ERG11 gene was upregulated in C. albicans isolates (23-25). It was proposed that the higher expression of this gene might contribute to the antifungal effect of fluconazole in C. albicans via the increased production of its target enzymes. According to the literature review, it seems that resistance to fluconazole is not only due to the changes in ERG11 but also some other mechanisms, such as the prevention of the intracellular accumulation of the antifungal agent and changes in the target enzyme, which lead to reduced drug attachment (26). The difference in the percentage of increased expression in different studies might be due to the synthesized method, concentration, and size of NPs. Song et al. showed that the administration of fluconazole had no significant effect on the expression rates of the ERG11 gene (27). Recently, Dižová revealed that fluconazole could cause a reduction in the expression level of the ERG11 gene only in combination with farnesol (28). In the current study, similar results were obtained, indicating that fluconazole only affects the expression level of this gene after being combined with ZnO-NPs. It might be proposed that the combined administration of fluconazole and ZnO-NPs might alter the biofilm formation via the regulation of the ERG11 gene.
It was proved that fluconazole causes decreased expression of the CDR2 gene alone and in combination with other drugs (29, 30). Shao et al. revealed that C. albicans isolates treated with fluconazole showed upregulation when compared to untreated control strains (31). The findings of the present study also demonstrated that the expression rate of CDR2 was upregulated in C. albicans isolates treated with fluconazole; nevertheless, its expression was interestingly downregulated when isolates were treated with ZnO-NPs and a combination of these two drugs. In addition, treating isolates with the combination of fluconazole and ZnO-NPs caused a much higher decrease in the expression level of this gene. This fact suggests that ZnO-NPs might facilitate the suppression of the efflux pumps in combination with fluconazole. Since the present study demonstrated the effect of this combination on the expression of several genes, it seems that resistance to fluconazole is multifactorial, involving other molecular mechanisms (26).
Differences in the gene expressions involved in C. albicans virulence might depend on the immunity system and mutations in the specific genes and the immunocompromised patients referring to clinics and hospitals frequently to receive antifungal drugs; therefore, the ERG11 gene expression level was changed (24). Golabek et al. indicated an increase in the expression of CDR2 and ERG11 in the azole-resistant strains of C. albicans in comparison to that of susceptible strains (32). Furthermore, they reported some ERG11 mutations which affect the expression of ERG11; consequently, Golabek et al. concluded that the mechanism of developing resistance to azoles might be a complex process. It seems that the evaluation of changes in the ERG11 and CDR2 sequences associated with gene expression in the local population can widely help understand the factors affecting the resistance of C. albicans to fluconazole.
The key step of Candida species adhesion to surfaces is biofilm formation, which causes persistent infection and microorganism attack on cells, particularly in hospitalized patients and individuals with the impaired immune system (33). As known, cell-surface-related glycosylphosphatidylinositol encoded by the ALS-family and HWP1 genes binds to glycoprotein, which mediates the attachment of C. albicans strains to mucosal surfaces. Functional analyses showed that the HWP1 gene and ALS1 gene have a key role in C. albicans biofilms, both at in vitro and in vivo levels. The role of these two genes in the colonization and virulence of C. albicans strains could be evaluated via the detection of their expression in C. albicans strains isolated from clinical specimens. Several studies (16, 34) indicated that, in comparison to fluconazole-sensitive isolates, their expression had a significant increase in fluconazole-resistant C. albicans isolates. The results of the present study showed that the treatment of C. albicans strains with the combination of ZnO-NPs and fluconazole in the tested isolates reduced the expression of the ALS1 and HWP1 genes significantly.
According to the literature review, it has been proposed that to trigger in vivo formation of biofilm, the HWP1 surface protein needs to be associated with the ALS gene family (35). Nas et al. reported that the expression of ALS1 and HWP1 genes was detected as 69% and 62% in all cases with vulvovaginal candidiasis (VVC), respectively. In pregnant, postmenopausal, and reproductive age women with VVC, the expression of the ALS1 gene was observed at 70%, 75%, and 67%, and the expression of the HWP1 gene was observed at 60%, 25%, and 73%, respectively (36). Finally, in line with the present study’s results, Hosseini et al. recently showed that the combination of fluconazole and ZnO-NPs caused a significant reduction in the expression levels of two ALS1 and ALS3 in comparison to those of the control strain (12). Some similar studies using different drugs in combination with fluconazole showed that the expression level of the HWP1 gene was significantly downregulated in C. albicans isolates (37, 38). Nevertheless, to the best of our knowledge, no study has examined the effect of ZnO-NPs on the expression rate of this gene. The current study showed that the treatment of C. albicans significantly reduced the expression of HWP1.
The main limitation of the present study was the low number of studied genes. There is a need for further comprehensive studies on other genes involved in biofilm formation in clinical isolates of C. albicans. Using a larger population with a higher number of isolated and a higher number of genes with a potential role in these pathways in future studies might lead to more reliable conclusions.
5.1. Conclusions
The findings of this study revealed that the simultaneous administration of ZnO-NPs and fluconazole could be more efficient in the inhibition of fungal growth and activity via decreasing the levels of ALS1, HWP1, and CDR2 gene expression. Therefore, ZnO-NPs eliminate the infection when combined with fluconazole or other antifungal agents. Based on the obtained results of this study and various tests of NPs, these ingredients are ideal for removing the infection in combination with new pharmaceutical formulations, along with antifungal chemical drugs conjugated with medications. As a result, further studies are required to investigate natural target cells on mouse models in vitro and in vivo.