1. Background
Bacterial and fungal plant diseases pose a major threat to agricultural production worldwide (1, 2). Nowadays, it remains a global challenge to develop new therapeutic modalities for treating infectious diseases caused by bacterial and fungal pathogens. A wide range of important secondary metabolites, including antibiotics and growth promoting substances, are produced by several members of Actinomycetes (3, 4). More than 60% of the nearly 6000 antibiotics of microbial origin are produced by Streptomyces spp. including both antibacterial and antifungal agents beside a considerable number of other bioactive compounds (5-11). Microorganisms represent a potentially valuable source of biologically significant compounds that should be explored for their potential agrochemical use (12). Soil strains of Actinomycetes are still important sources of novel antibiotics.
In the past decades, although many species, which produce biologically active metabolites, have been obtained from soil samples, the chance of isolating a new Actinomycete strain from a common terrestrial habitant has reduced markedly. In the process of screening new agricultural antibiotics, researchers have to look for novel microorganisms in unusual environments. Chemically polluted soil is one sort of such unusual environments. Chemical pollutants could be mutagens, and some of the mutant strains might give rise to increased productivity of bioactive metabolites, or even produce new bioactive compounds (13, 14).
2. Objectives
In this study, we report the compounds ZM-1 and ZM-2 of the fermentation broth of Streptomyces parvus 33, which was isolated from the chemically polluted soil samples. The compound ZM-1 was isolated in the form of a clear crystal for the first time.
3. Methods
3.1. General Materials and Selected Microorganism
Solvents were of analytical grade (AR) unless otherwise mentioned. TLC was performed on 60 F254 silica gel plates (Merck, USA). Column chromatography was used on HPD100 macroporous resin (Baoen, China) with methanol elution (MeOH, qingdao marine chemical, China). A Waters 2692 HPLC apparatus (Waters, USA) equipped with a Sinochrom ODS-BP (10 × 300 mm, 10 µm, Shimadzu, Japan) reverse phase column was employed using methanol-water as mobile phase at flow rate of 3.0 mL/min, monitored by UV detector at 240 nm.
The selected microorganism: Pathogenic bacteria were Solanacearum (Ralstonia solanacearum), Pseudomonas syringae pv. Actinidiae provided by the college of plant protection, Northwest A and F University. Bacillus cereus (Waxy bacillus, 1.184), B. subtilis (Bair conditionerillus subtilis, 1.88), P. aeruginosa (1.2031), Staphylococcus aureus (1.89), and Escherichia coli (1.1574) were purchased from China general microbiological culture collection center. The pathogenic bacteria were inoculated in beef extract peptone agar culture medium at 37°C for 1 day. Pathogenic fungi were Fusarium graminerum, Curvularia lunata, Colletotrichum orbiculare, C. gloesporioides provided by the College of Plant Protection, Northwest A & F University. The inoculation of the pathogenic fungi was carried out in potato dextrose agar (PDA) medium at 25°C for 3 - 7 days.
3.2. Isolation and Identification of Streptomyces parvus 33
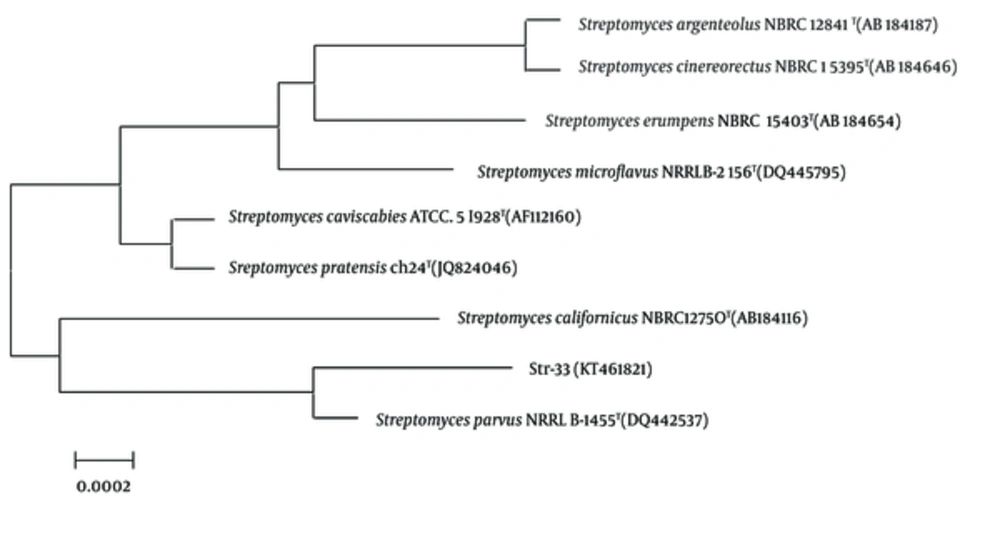
The strain 33 was isolated from a chemically polluted soil sample collected from the Shaanxi province of China. According to the results of morphological examination and culture characteristics, physiology and biochemical measurement and 16S rDNA sequence analysis, the strain 33 was highly homological (up to 99%) with S. parvus NRRLB- 1455T (Figure 1). Therefore, the strain 33 was identified and designated as S. parvus 33. The voucher specimen of the Streptomycete was deposited in the Microbiology Institute of Shaanxi, China.
Streptomyces parvus 33 was cultivated at 28°C in Gause’s medium, which contained soluble starch (2%), K2HPO4 (0.05%), KNO3 (0.1%), NaCl (0.05%), MgSO4•7H2O (0.05%), FeSO4•7H2O (0.001%), and agar (1.5%), pH = 7.2 - 7.4. Fermentation was performed in two stages: seed growth and production. The spores of strain 33 grown on Gause’s medium were used to inoculate a 250 mL flask containing 60 mL of a sterile seed medium consisting of glucose (1.0%), millet steep liquor (1.0%), peptone (0.5%), (NH4)2SO4 (0.1%), NaCl (0.25%), and CaCO3 (0.05%), pH 7.2. The flask was shaken on a shaker at 180 rpm for 18 h at 28°C. 6 mL of the seed culture were transferred to 250 mL flasks containing 60 mL of a sterile production medium consisting of glucose (1.0%), millet steep liquor (1.0%), peptone (0.3%), (NH4)2SO4 (0.1%), NaCl (0.25%), and CaCO3 (0.1%), pH 7.2. Fermentation was carried out at 180 rpm for 4 days at 28°C on a rotary shaker.
3.3. Extraction and Isolation
The culture of 90 L of S. parvus 33 was filtered through cheesecloth to separate the medium and culture liquid at 25°C, pH 7.0. The filtrate was absorbed onto HPD-100 macroporous resin, and then eluted with MeOH. The MeOH fraction was evaporated in vacuum. The concentrate was subjected to column chromatography and eluted with petroleum ether and EtOAc, in sequence. The antimicrobial fraction was concentrated under vacuum, and further purified on a Waters 2695 HPLC apparatus equipped with a Sinochrom ODS-BP (10.0 mm × 300 mm, 10 μm) reverse phase column, using 50% MeOH/H2O as the mobile phase at flow rate of 3.0 mL/min, monitored by UV detector at 240 nm to afford two compounds ZM-1 and ZM-2. ZM-1 was obtained as a single crystal, and determined by single crystal diffraction. ZM-2 was obtained as colorless amorphous solid.
3.4. Antimicrobial Activity
3.4.1 Inhibition of Bacteria
Minimum inhibitory concentration (MIC) was (15) determined against bacteria using micro-dilution method (Method 96): 5 mL sterile MH broth was added to a test tube; using an inoculating needle, a small amount of bacterial broth was picked and added to the tube, placed in 37°C incubator for 12 hours. The absorbance of cell culture was measured using UV-Vis spectrophotometer at 600 nm against a culture medium without bacteria as blank control, according to each equivalent to a concentration of 0.1 OD600 1 × 108 cfu/mL, were diluted to a concentration of 1 × 106 cfu/mL.
To conduct tests with 96-well culture plates (12 × 8, U-shaped hole at the end), 0.1 mg of the test compound was first dissolved in 5 μL dimethyl sulfoxide (DMSO), and then dissolved in 100 mL sterile water to be formulated at a concentration of 100 μg/mL as the mother liquor. The first test well was filled with 50 μL maximum concentration of the drug solution, each of 50 μL sterile water are added, the double dilution method using a gradient of 1 - 8 hole diluted to 500 ~ 3.9 μg/mL gradient series. The 9th and 10th holes were filled with 50 μL sterile water as negative control. Then, it was added at a concentration of 1 to 9 holes 1 × 106 cfu/mL of bacteria solution 50 μL, 10th hole by adding a blank incubation 50 μL sterile water. The 11th hole was filled with 50 μL 2.5% DMSO and 50 μL solution at a concentration of 1 × 106 cfu/mL of the broth. The 12th hole was filled with 100 μL sterile water as blank. Then, the microporous oscillation mixing was performed in 37°C incubator while ampicillin was used as positive control. Each treatment was repeated three times.
12 hours after treatment, the results were observed: in the case of a black background light observation, there are holes in the bottom of the growth of bacteria precipitate or diffuse type turbidity, the lowest sample concentration of holes contained sterile growth inhibition is the lowest of the sample bacterial concentration (MIC).
3.4.2. Inhibition of Plant Pathogenic Fungi
Using suppression spore germination: pathogenic fungi spores will develop well paired with sterile water at a concentration of 1 × 105 ~ 1 × 107 spore suspension. 0.1 mg of the test compound was first dissolved in 10 μL DMSO, and then dissolved in 200 mL sterile water, formulated at a concentration of 50 μg/mL of the mother liquor (16). The agent activity was set at final concentrations of 25 μg/mL, 12.5 μg/mL, 6.25 μg/mL, as 3 liquid concentration gradients.
With a pipette to draw 25 μL and 25 μL each concentration liquid droplets prepared spore suspension was added to the dry cleaner’s concave slide, so that the liquid and mix equal amounts of spore suspension, then placed with shallow water dish, capped at 25°C constant moisture incubator. Each treatment was repeated three times against distilled water. When the control spore germination rate was more than 85%, spores were checked in each treatment (per treatment were observed each repeated five horizons, the total number of spores survey less than 250) germination under an optical microscope. Table 2 shows a formula agent for inhibition rate of spore germination.

4. Results
4.1. Chemistry
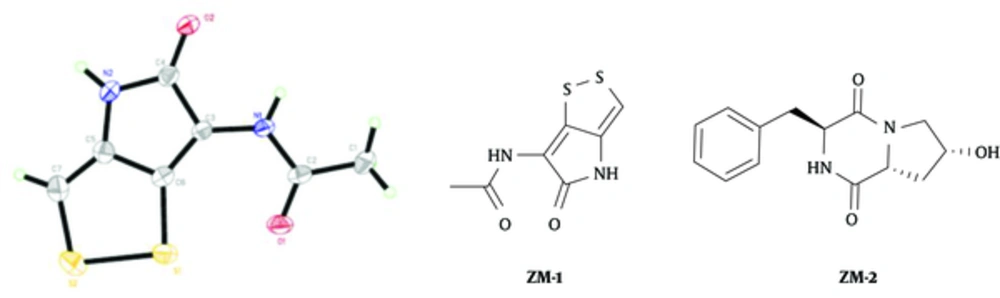
The active ingredients were obtained by the techniques of macroporous resin adsorption, silica gel column chromatography, and preparative reverse phase high performance liquid chromatography. The compound ZM-1 was obtained in the form of a clear single crystal, and unambiguously confirmed by X-ray crystallography. ZM-2 was characterized by IR, 1H-NMR, 13C-NMR, DEPT (90° and 135°), and MS analysis, as illustrated in Figure 2.
The compound ZM-2: colorless amorphous solid; HR-ESI-MS (positive) m/z: 283.1059 [M + Na]+ (calcd for C14H16N2O3Na, 283.1059). 1HNMR (400 MHz, CD3OD) δ: 7.18 - 7.32 (5H, m, H-12-H-16), 4.22 (2H, m, H-3 and H-8), 3.87 (1H, dd, J = 12.4,3.9 Hz, H-9a), 3.27 (1H, dd, J = 12.6, 4.9Hz, H-9b), 3.19 (1H, dd, J = 5.1, 13.6 Hz, H-10a), 3.00 (1H, dd, J = 4.8, 13.6 Hz, H-10b), 2.78 (1H, t, J = 8.5 Hz, H-6), 2.23 (1H, m, H-7a), and 1.92 (1H,m, H-7b); 13C NMR (CD3OD) δ: 170.98 (C-5), 167.70 (C-2),136.78 (C-11), 131.20 (C-12 and C-16), 129.70 (C-13 and C-15), 128.53 (C-14), 68.47 (C-8), 59.61 (C-3), 57.20 (C-6), 53.99 (C-9), 40.86 (C-10), and 38.0 (C-7).
4.2. Biological Assay
The antimicrobial activity of ZM-1 and ZM-2 was measured according to a previously reported method. The biological activities of ZM-1 and ZM-2 towards a wide variety of bacterial and fungal plant pathogens were evaluated, giving the following results. The inhibitory effects of ZM-1 against bacteria are listed in Table 1. Antibacterial activities were measured by the micro-broth dilution method in 96-well culture plates, with Ampicillin as a positive control, to evaluate the biological activities of the compound ZM-1 against the tested bacteria at different concentrations. For most of the tested strains, the minimum inhibitory concentration of the compound ZM-1 was 0.39 μg/mL, which is less than that of the positive control ampicillin. Especially for the P. syringae pv. Actinidiae and the R. solanacearum, the value of MIC of the positive control was 160 times more than that of the compound ZM-1, which shows that the antibacterial ability of the compound ZM-1 is strong.
| Tested Bacteria | Minimum Inhibitory Concentration MIC (μg/mL) | |
|---|---|---|
| ZM-1 | Ampicillin | |
| Bacillus cereus | 0.78 | 3.13 |
| Bacillus subtilis | 0.39 | 12.5 |
| Staphylococcus aureus | 0.39 | 6.25 |
| Escherichia coil | 0.39 | 25 |
| Pseudomonas aeruginosa | > 50 | 25 |
| Pseuomonassyringaepv. Actinidiae | 0.39 | > 50 |
| Ralstoniasolanacearum | 0.39 | > 50 |
The inhibitory effects of ZM-1 against spore germination are listed in Table 2. Spore germination inhibition assay was utilized to investigate the biological activities of ZM-1. The compound ZM-1 exhibited a high degree of activity against the tested pathogenic fungi. The antimicrobial activity was enhanced with the increase of concentration of the compound ZM-1. However, ZM-2 was not effective against the tested bacterial and fungal plant pathogens.
| Tested Pathogenic Fungi | Inhibition Rate of Spore Germination (%) | ||
|---|---|---|---|
| 6.25μg/mL | 12.5μg/mL | 25μg/mL | |
| Fusariumoxysporum f sp. vasinfectum | 25.93 ± 0.17 | 69.14 ± 0.34 | 93.98 ± 0.54 |
| Curvularialunata | 24.44 ± 1.39 | 39.87 ± 0.80 | 94.97 ± 1.17 |
| Colletotrichumorbiculare | 23.36 ± 0.73 | 55.64 ± 1.05 | 96.19 ± 0.45 |
| Colletotrichumgloesporioides | 37.81 ± 0.56 | 65.73 ± 1.09 | 89.85 ± 1.21 |
The compounds ZM-1 and ZM-2 were isolated from the extract of the fermented broth of S. parvus 33 by bioassay-guided fractionation. The compound ZM-1 was isolated in the form of a clear single crystal for the first time, and identified as holomycin via X-ray crystallography. Streptomyces parvus 33 is a newly discovered producer of holomycin. ZM-1 showed a strong antibacterial activity against the tested bacteria, such as B. cereus, B. subtilis, E. coli, S. aureus, and P. syringaepv. actinidiae, and its value of MIC was greater than that of the positive control ampicillin. The compound ZM-1 exhibited a high degree of activity against F. oxysporumf. sp. vasinfectum, C. lunata, C. orbiculare, and C. gloesporioides. The antimicrobial activities were enhanced with the increased concentration of the compound ZM-1. However, ZM-2 was not effective against the tested bacterial and fungal plant pathogens. In general, the compound ZM-1 is a valuable lead compound for the development of agricultural fungicides while acts against bacteria, as well.