1. Background
Chronic ulcers do not improve within 1 - 3 months (1). In such ulcers, several factors usually delay the ulcer healing process. These factors include endocrine disorders (diabetes), vascular problems, immune system dysfunction, infection, and bacterial biofilm formation (2). The most important chronic ulcers encompass diabetic foot ulcers, venous leg ulcers, pressure ulcers, and surgical site infections (3). Ulcer healing has four successive stages, including homeostasis, inflammation, proliferation, and regeneration, and chronic ulcers often remain in the inflammation stage (4). Studies indicate that 70% of chronic ulcers have biofilms, while only 6% of acute ulcers, which have timely healing, form biofilms (5). Bacterial resistance to antimicrobials in the biofilm phase is a major global issue (6, 7). By providing special conditions, biofilms enable the horizontal transfer of antibiotic-resistance genes and virulence factors among bacteria, thereby minimizing the effectiveness of antibiotics and antiseptics and causing the overuse of antibiotics and the spread of multiple-antibiotic resistance. Hence, reducing the bacterial load is a key parameter in the ulcer healing path (8).
Many studies have examined the antimicrobial activity of probiotics through the production of bacteriocins, lactic acid, hydrogen peroxide, and other antimicrobial molecules (9). In this regard, Lactobacillus lactis has a special place due to the production of nisin bacteriocin, and several studies have demonstrated its antibacterial function (10). Lactobacillus delbrueckii also competes with pathogenic bacteria by producing lactic acid and propionic acid and decreasing pH, thus inhibiting their growth (11). It is worth noting that bacteria in the biofilm exhibit high resistance to antimicrobial agents compared to the planktonic form; hence, it is necessary to consider the physiological status of the ulcer and the presence of bacteria in the biofilm to evaluate the inhibitory effect of probiotics. Besser et al. overcame this challenge by designing a novel in vivo like human plasma biofilm model (hpBIOM) and studied the anti-biofilm activity of L. plantarum, Bifidobacterium lactis, and Saccharomyces cerevisiae on clinically isolated biofilms (12). The hpBIOM-aided evaluation of the anti-biofilm potential of other probiotics provides valuable information for the biofilm treatment of chronic ulcers by bacteriotherapy.
2. Objectives
Since there was no information about the effects of two probiotics, L. lactis, and L. delbrueckii, on inhibiting and destroying biofilms of chronic ulcers separately and simultaneously, the present study examined the simultaneous effects of the two probiotics on the inhibition of pathogens isolated from chronic ulcer biofilms with multiple-drug resistance (MDR) in hpBIOM.
3. Methods
3.1. Specimen Collection
From July 2020 to February 2021, 82 specimens of chronic ulcer biofilms were collected from patients who visited the Khanevadeh and Al-Zahra Hospitals in Isfahan. Inclusion criteria were as follows: The presence of chronic ulcers lacking timely healing, the presence of biofilm in the ulcer, and consumption of at least a period of antibiotics. Ulcers were first cleaned with sterile normal saline; then, biopsy specimens were obtained from ulcer biofilms with forceps and transferred to a laboratory in a nutrient transfer medium for microbial examination.
3.2. Isolation and Identification of Bacterial Isolates
The specimens were cultured on MacConkey and blood agar (QuE-Lab, Italy) and incubated at 37°C for 24 hours. After isolation, the isolates were identified using Gram staining and conventional diagnostic tests. Finally, a universal primer was used to amplify the 16S rRNA gene for molecular identification, and the PCR product was sent to Zist Exir Company for sequencing (13).
3.3. Antibiotic Susceptibility of Isolates
The antibiotic resistance pattern of the isolates was determined using the Kirby-Bauer method according to CLSI standards for the following antibiotics: Imipenem (10 µg), amoxicillin-clavulanic acid (10/20 µg), ciprofloxacin (5 µg), cefoxitin (30 µg), ceftriaxone (30 µg), cefepime (30 µg), cefotaxime (30 µg), Co-trimoxazole (1.25/23.75 µg), penicillin (10 µg), clindamycin (2 µg), gentamicin (10 µg), and ampicillin (10 µg) (BD BBL, Canada). The susceptibility of the isolates to colistin and vancomycin was examined using E-test, and the corresponding minimum inhibitory concentration (MIC) values were determined. After determining the antibiotic resistance pattern, three isolates, namely Pseudomonas aeruginosa, Staphylococcus aureus, and Klebsiella pneumoniae, with multiple-antibiotic resistance, were selected for further analysis.
3.4. Preparation of Probiotics
Two probiotics, L. lactis, and L. delbrueckii, were isolated from the industrial yogurt starter of Hansen Company. After confirmation of phenotypic identity using biochemical tests, the probiotics were separately cultured in MRS broth at 37°C and 100 - 150 rpm in a shaker incubator for 48 hours.
3.5. Human Plasma Biofilm Model (hpBIOM) Preparation
The hpBIOM was prepared based on the method proposed by Besser et al. (14). Plasma and buffy coats were collected from donors. First, the buffy coat was centrifuged at room temperature and 3,000 rpm for 30 minutes to remove residual red blood cells. The platelet-rich plasma and buffer coat were then fused and shaken continuously in a test tube at 100 rpm at 22°C. For 1 mL of plasma, 1 mL of the bacterial isolate was added at a concentration of 0.5 McFarland (1.5*108), and then 18.27 μL of CaCl2 was added, mixed well, and quickly transferred to 12-well culture plates. The plates were incubated for two hours at 37°C and 50 rpm to form polymerized plasma and disks of bacterial biofilm. After preparing the biofilm disk, 100 μL of suspension of each probiotic with a turbidity of 109 CFU was added to the top of the disk, and the second dose of probiotic was added with the same amount and turbidity after two hours and was then incubated at 37°C. To investigate the simultaneous effects of L. lactis and L. delbrueckii on the biofilms of isolates, 50 μL of each probiotic with a turbidity of 109 CFU was added to the disk in two doses with an interval of two hours.
3.6. Biofilm Dissolution and Bacterial Growth Count
For enzymatic digestion of plasma, 1 mL of bromelain (b4882 Sigma-Aldrich) at a concentration of 0.1 g/mL in PBS was added to the wells two and 24 hours after adding the second dose of probiotic, followed by incubation at 37°C for two hours. To count the surviving pathogenic bacterial cells, serial dilutions were prepared from 100 μL of solution of each well and inoculated on MRS TSA agar. Then, the plates were incubated at 37°C overnight. Finally, colony counting was performed.
3.7. Field Emission-Scanning Electron Microscopy (FE-SEM)
In the present study, the interaction of L. delbrueckii probiotic with P. aeruginosa isolates and the simultaneous interaction of L. delbrueckii and L. lactis with P. aeruginosa after 24 hours were investigated using field emission-scanning electron microscopy (FE-SEM).
3.8. Statistical Analysis
Data were expressed as mean ± SD. The experiments were performed in triplicate to evaluate the anti-biofilm activity of probiotics for each plasma donor. The Mann-Whitney test was used to compare the results between treatment and control groups, and P < 0.05 was considered significant. GraphPad PrismTM software version 9 was used.
4. Results
The present descriptive cross-sectional study examined 82 patients, including 56 (68.3%) men and 26 (31.7%) women with chronic ulcers who visited Khandevadeh and Al-Zahra Hospitals in Isfahan. Among them, the frequency of bedsores was 18.3%, diabetic ulcers 63.4%, arterial ulcers 6.1%, and venous ulcers 12.2%. Most patients (41.5%) were in the age range of 61 - 70 years. The duration of ulcer involvement was two to three months in most patients (31.7%). According to Fisher's exact test results, the frequency distribution of the cause of ulcers was not statistically different between different age groups (P = 0.986).
4.1. Bacterial Isolates
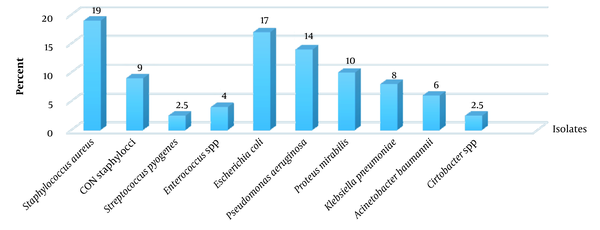
Among 82 biofilm specimens, 47 (57.3%) and 35 (42.7%) were monomicrobial and polymicrobial, respectively. There were 33 specimens containing two bacteria, and two specimens had three bacteria. The frequency of Gram-negative and Gram-positive isolates was 65.5% and 34.5%, respectively. Among Gram-negative bacteria, Escherichia coli (17%), P. aeruginosa (14%), Proteus mirabilis (10%), K. pneumoniae (8%), and Acinetobacter baumannii (6%) were predominant. Staphylococcus species (80.5%) were the predominant pathogen among Gram-positive bacteria. Staphylococcus aureus (19%) and coagulase-negative staphylococci (9%) were the predominant pathogens among Gram-positive bacteria (Figure 1).
4.2. Antibiotic Resistance Patterns of Isolates
Table 1 presents the antimicrobial resistance patterns of Gram-positive and Gram-negative isolates. All staphylococcal isolates were resistant to penicillin, of which 27% were methicillin-resistant S. aureus (MRSA), and resistance to ciprofloxacin was 63.6%; also, vancomycin was the most effective antibiotic against Gram-positive bacteria. The highest resistance to imipenem was observed in E. coli isolates (75.0%). The percentage of colistin resistance was 29.5 % in P. aeruginosa. Among Gram-negative isolates, K. pneumoniae had high resistance to cefotaxime (66.6%), ceftriaxone (55.5%), colistin (33.3 %), and co-trimoxazole (77.7%). On the contrary, the most effective antibiotics against Gram-negative bacteria were cefepime and cefoxitin. In general, 59.9% of the isolates were MDR. The MDR percentages among Gram-positive and Gram-negative isolates were 61% and 59%, respectively.
| Type of Antibiotic | S. aureus, N = 22 | CON Staphylococci, N = 11 | E. coli, N = 20 | P. aeruginosa, N = 17 | P. mirabilis, N = 12 | K. pneumonia, N = 9 | A. baumannii, N = 7 |
|---|---|---|---|---|---|---|---|
| PEN | 22 (100.0) | 11 (100.0) | NT | NT | NT | NT | NT |
| CLI | 13 (59) | 7 (63.6) | NT | NT | NT | NT | NT |
| VAN | (0.0) | (0.0) | NT | NT | NT | NT | NT |
| AMC | 5 (23) | 4 (36.3) | 3 (15.0) | 5 (29.5) | 2 (16.6) | 2 (22.2) | 2 (28.5) |
| FEP | NT | NT | 3 (15.0) | 1 (5.8) | 2 (16.6) | 2 (22.2) | 1 (14.3) |
| CTX | NT | NT | 7 (35.0) | NT | 5 (41.6) | 6 (66.6) | NT |
| FOX | 6 (27) | 3 (27) | 3 (15.0) | NT | 1 (8.3 ) | 1( 11.1) | NT |
| CRO | NT | NT | 7 (35.0) | 9 (53.0) | 6 (50.0) | 5 (55.5) | 3 (43.0) |
| CIP | 14 (63.6) | 7 (63.6) | 14 (70) | 11 (64.7) | 9 (75.0) | 7 (77.7) | 5 (71.4) |
| CST | NT | NT | 4 (20.0) | 5 (29.5 ) | 2 (16.6 ) | 3 (33.3 ) | 1 (14.3) |
| GEN | 5 (23) | 2 (18.2) | 6 (30.0) | 11 (64.7) | 6 (50.0) | 5 (55.5) | 3 (43.0 ) |
| IPM | NT | NT | 15 (75.0) | 13 (76.4) | 8 (66.6) | 8 (88.8) | 5 (71.4 ) |
| SXT | 4 (18.2) | 3 (27) | 12 (60.0) | 10 (59.0) | 8 (66.6) | 7 (77.7) | 3 (43.0 ) |
| AMP | NT | NT | 13 (65.0) | NT | 8 (66.6) | 6 (66.6) | NT |
Antibiotic Resistance Patterns of the Most Frequent Bacterial Isolates from Chronic Ulcer Biofilm a
4.3. Interaction of Lactobacillus delbrueckii with Biofilms of MDR Isolates
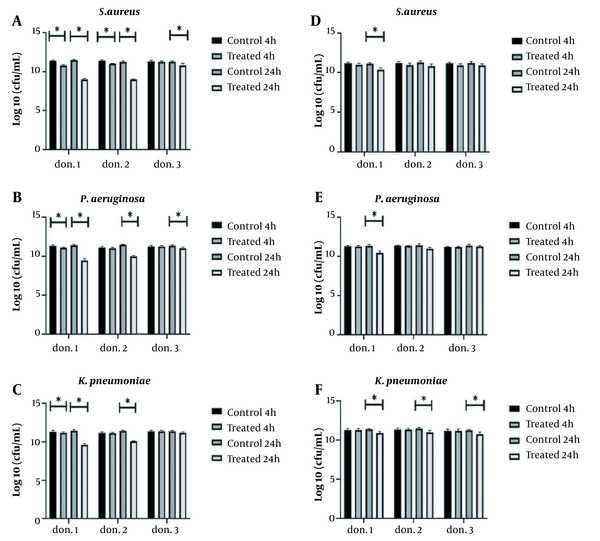
The study of the anti-biofilm activity of L. delbrueckii against three MDR pathogens, S. aureus, P. aeruginosa, and K. pneumoniae, indicated the significant effect of this probiotic against all three pathogens, but L. delbrueckii could not completely remove pathogenic isolates. However, it significantly inhibited bacterial growth in plasma biofilm over four hours against three pathogens in donor 1 (P < 0.05) (Figure 2A-C). In donors 1 and 2, significant anti-biofilm activity was observed against S. aureus during 24 hours, and it was accompanied by an average log10 reduction of 2.36 CFU/mL. The same result was also observed for P. aeruginosa, and a log10 reduction of 1.43 CFU/mL was detected within 24 hours (Figure 2A and B). In donor 3, no decrease in K. pneumoniae biofilm was observed by L. delbrueckii; however, there was a significant decrease in donor 1, after four and 24 hours, compared to the control group (P < 0.05). The antibacterial activity of L. delbrueckii was higher in donor 1 than in the other two donors (Figure 2C).
4.4. Interaction of Lactobacillus lactis with the Biofilm of Pathogenic Isolates
Lactobacillus lactis resulted in a significant reduction in the growth of the three pathogens after 24 hours of incubation in donor 1. The antimicrobial activity of L. lactis against S. aureus and P. aeruginosa had the same pattern. That means no change was observed in hpBIOMs in donors 2 and 3, and a log10 reduction of 0.74 - 0.88 CFU/mL was detected in the biofilm of pathogens in donor 1. In the case of K. pneumoniae, a log10 reduction of 0.47 CFU/mL was observed in all three donors after 24 hours (Figure 3D-F).
Antimicrobial activity of Lactobacillus delbrueckii and L. lactis in hpBIOM. A, B and C, L. delbrueckii significantly inhibited the growth of Staphylococcus aureus and Pseudomonas aeruginosa in all three donors within 24 hours but did not affect Klebsiella pneumoniae biofilm in the third donor. L. delbrueckii showed the best anti-biofilm activity in the first donor plasma; D, E and F, antimicrobial activity of L. lactis against S. aureus and P. aeruginosa, except for K. pneumoniae, was donor-plasma dependent. All tests were repeated three times (*: P < 0.05).
4.5. Simultaneous Effects of Lactobacillus lactis and Lactobacillus delbrueckii on Biofilm in hpBIOM
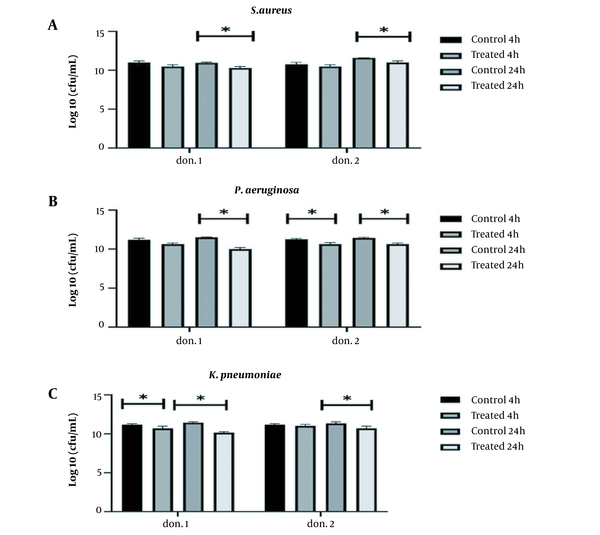
The present study examined the simultaneous interaction of two probiotics, L. lactis, and L. delbrueckii, against three MDR isolated from clinical specimens using donor plasma, with better antibiofilm activity. Two probiotics simultaneously prevented the formation of biofilms by all three pathogens within 24 hours compared to the control group (P < 0.05) (Figure 2). The highest anti-biofilm activity of two probiotics was simultaneously observed on P. aeruginosa with a log10 reduction at 1.47 CFU/mL and 0.83 CFU/mL in the first and second donors, respectively. However, comparing the inhibitory effects of the two probiotics simultaneously and alone demonstrated that they did not synergize the antibiofilm activity of each other.
4.6. Findings of FE-SEM
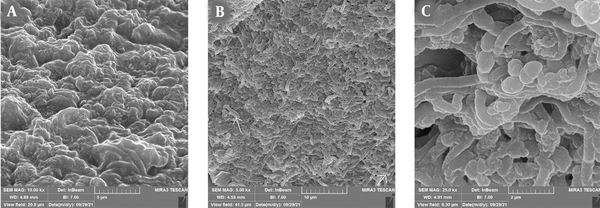
Field emission-scanning electron microscopy (FE-SEM) was used for a more accurate analysis of the anti-biofilm activity of L. delbrueckii on P. aeruginosa. A comparison of control and treatment images revealed the influence of L. delbrueckii on hpBIOM. The number of P. aeruginosa cells was significantly reduced, and the matrix was covered largely by L. delbrueckii (Figure 4A and B). The FE-SEM image of the simultaneous interaction of two probiotics indicated the formation of exopolysaccharides by L. delbrueckii, among which L. lactis was obvious and replaced P. aeruginosa cells (Figure 4C).
FE-SEM images of simultaneous effects of Lactobacillus lactis and L. delbrueckii on the growth of Pseudomonas aeruginosa in hpBIOM: A, P. aeruginosa biofilm after 24 hours that is associated with high production of exopolysaccharide; B, P. aeruginosa after exposure to L. delbrueckii, the surface of which is covered with microcolonies containing probiotics and a very small number of P. aeruginosa (arrow); C, P. aeruginosa biofilm in simultaneous exposure to L. lactis and L. delbrueckii, where a high density of two probiotics was visible.
5. Discussion
Chronic ulcers have become a major challenge worldwide by increasing economic costs and impacting patients' quality of life (15). In the present study, similar to other studies, the most common bacteria isolated from chronic wounds were Gram-negative bacteria belonging to the Enterobacteriaceae family (16). Among them, E. coli was the most common Gram-negative bacterium. In contrast, Wong et al. reported P. aeruginosa as the most common Gram-negative bacterium in chronic wounds (17). The frequency of MRSA strains in our study was consistent with Ralph's findings (18). In the present study, like Silva et al., the frequency of MDR bacterial isolates was high (19). Based on the findings of this study, the resistance of Gram-negative isolates to fluoroquinolones (ciprofloxacin) and carbapenem (imipenem) was increasing.
The biofilm formation by bacteria is a non-endogenous reason making chronic ulcer healing difficult. Bacteria in polysaccharides of the biofilm matrix have phenotypic resistance compared to the planktonic form and genetically acquire their resistance to antimicrobials by horizontal transfer of resistance genes in the biofilm (20). This study examined the anti-biofilm activity of two probiotics, L. lactis, and L. delbrueckii, on three MDR pathogens isolated from chronic ulcers in hpBIOM. This study used this model since hpBIOM largely mimics the human ulcer biofilm. In the present study, L. delbrueckii showed significant anti-biofilm activity against the three pathogens during 24 hours of incubation. Lactobacillus supernatant contains exopolysaccharides and biosurfactants that inhibit bacterial biofilms (21, 22). Kim et al. reported that L. acidophilus exopolysaccharides could inhibit E. coli enterohemorrhagic biofilm in the microplate. It was also reported that the biosurfactants produced by Lactobacillus RC-14 considerably reduced S. aureus attachment to surgical implants (23).
The cell-free supernatant and whole-cell of L. delbrueckii had stronger antimicrobial activity on S. aureus than on P. aeruginosa and K. pneumoniae. Gram-negative bacteria are more resistant to antimicrobial compounds in supernatants due to an outer membrane in their wall (24). Furthermore, P. aeruginosa can develop resistance to antimicrobial agents in the cell-free supernatant. Pseudomonas aeruginosa can form a dense biofilm that increases its resistance to antimicrobials in the supernatant. In the present study, L. lactis exhibited a lower antimicrobial effect than L. delbrueckii. Nisin bacteriocin is an antimicrobial compound produced by L. lactis and has strong antimicrobial effects against Gram-positive bacteria (25). However, nisin-like bacteriocins are also produced by some L. lactis strains that affect Gram-negative pathogens, including Salmonella typhimurium. Kuwano et al. reported that nisin Z made the cytoplasmic membrane of Gram-negative and Gram-positive bacteria permeable (26). The low anti-biofilm activity of L. lactis in this study was probably due to a lack of optimal production of nisin since its production was influenced by environmental factors, such as pH, incubation temperature, and carbon source (27, 28).
Unlike Besser's study, none of the probiotics could eliminate the pathogens (12). The difference was probably due to the types of probiotics used and the MDR of pathogens. The MDR bacteria use resistance mechanisms like efflux pumps that play a crucial role in their resistance to antimicrobial agents in the supernatant (29). The results of FE-SEM studies indicated that probiotics in hpBIOM in competition with pathogenic species prevented the biofilm formation by pathogenic bacteria through competitive removal, spatial inhibition, and colonization. Chan et al. found that the lipoteichoic acid (LTA) in the cell wall of Lactobacillus prevented the binding of uropathogenic bacteria to epithelial cells (29).
For the first time, this study examined the simultaneous effect of the two probiotics on the biofilm of MDR pathogens in hpBIOM. The results demonstrated the non-synergistic effects of L. lactis and L. delbrueckii bacteria on the biofilm of pathogens. The anti-biofilm activity decreased compared to the situation in which only a probiotic was used. According to studies, synergistic interactions often occur between probiotics when there is a common metabolite between the two probiotics. Somkuti and Steinberg investigated the synergistic effect on pediocin production by L. delbrueckii. Lactobacillus delbrueckii metabolizes lactose to glucose and galactose by producing β-galactosidase in the medium and provides the substrate for Pediococcus acidilactici (30).
Another study indicated a synergistic interaction between lactic acid bacteria (LAB) and Saccharomyces cerevisiae. Yeast increased the growth of L. sanfranciscensis by producing the growth factor and carbon dioxide in the dough, but the growth of L. delbrueckii by yeast depended on the carbon source in the dough. In the present study, the reduction of anti-biofilm activity in the simultaneous presence of two probiotics was probably due to the competition between the two types of probiotics for food and, consequently, a decrease in the production of antimicrobial agents. The donor-specific antibacterial activity of probiotics was a significant finding of the present study, which was consistent with Besser's study. The finding highlights the need to investigate donors' immunological characteristics and their effect on the anti-biofilm activity of LAB in future studies.
5.1. Conclusions
This study showed that L. delbrueckii and L. lactis have antibacterial activity against the biofilms of MDR isolates from chronic wound infection. However, their synergistic effects on these isolates' biofilm were insignificant. Therefore, more research is necessary to find probiotics with significant synergistic effects. Since the anti-biofilm activity of probiotics in this model was affected by donor plasma, conducting detailed immunological studies in this field indicates the prospect of using probiotics in vivo.