Abstract
Background:
Azole resistance rates are rising in Candida species. Fluconazole is one of the most important antifungal drugs used in candidiasis treatment.Objectives:
We identified the molecular mechanisms of fluconazole resistance of Candida albicans oropharyngeal candidiasis (OPC) isolates obtained from head and neck cancer patients, a study carried out between 2018 and 2020.Methods:
One hundred and twenty-five C. albicans clinical isolates were collected. Antifungal susceptibilities were determined by the CLSI- M27-A3 method. The ERG11 gene was amplified and sequenced to discover SNP mutation. Moreover, real-time PCR was carried out to measure the mRNA levels of ERG11, CDR1, CDR2, and MDR1.Results:
Resistance to fluconazole was found in 15 C. albicans isolates. Amino acid substitutions E266D and D116E were observed in resistant, sensitive dose-dependent (SDD), and susceptible C. albicans isolates. K128T, G465S, A114S, Y257H and V488I were in relation to fluconazole resistance. D504A, P375A, W520C, G59S, and V51L were novel substitutions detected in the isolates; except for D504A, other mutations were observed only in resistance isolates. The expression levels of CDR2, CDR1, MDR1, and ERG11 were increased compared to susceptible isolates, respectively.Conclusions:
ERG11 mutation was the principal mechanism for fluconazole resistance in C. albicans isolated from oropharyngeal candidiasis patients, and caspofungin can be used as the effective antifungal substance in fluconazole resistance situation for C. albicans infection.Keywords
Candida albicans Oropharyngeal Candidiasis Azole-resistance Gene
1. Background
Among human fungal diseases, Candida albicans is considered the most common opportunistic pathogen, which causes various illnesses ranging from superficial to life-threatening conditions (1-3). Although C. albicans may be harmless due to the balance kept with other existing microbiota in healthy individuals, it can immediately turn into a pathogen under certain circumstances (4, 5). In other words, alterations in the immune system and variations in host health can result in enabling these pathogens to cause infection (6, 7). It has been revealed that candidiasis is a fungal infection in which C. albicans is considered the most prevalent causative agent (8). Radiation therapy on head and neck cancer patients enhances the risk of developing candidiasis. This condition leads to oropharyngeal candidiasis (OPC) (9, 10), which is a common oral complication causing systemic infection in vulnerable hosts (11).
Significant research has been done on antifungal drugs to provide treatment for systemic yeast infections. These compounds with therapeutic properties include four main classes: azoles (fluconazole, itraconazole, voriconazole, posaconazole), polyenes (amphotericin B), echinocandins (caspofungin, micafungin, anidulafungin, aminocandin), and antimetabolites (5-flourocytosine) (12, 13). Fluconazole, a bis-triazole antifungal agent, is thought to as the first-line cure for systemic infection of Candida (14, 15). Fluconazole’s function is to hamper lanosterol C-14-a-demethylase enzyme, which is required for converting lanosterol to ergosterol. Subsequently, aggregation of 14-a-methyl sterol and drop in ergosterol in yeast cell wall leads to prevention of cell proliferation (16).
As aforementioned, the extensive long-term administration of agents with fungicidal activity, especially azoles, has led to the emergence of a variety of species resistant to these drugs (17). Several classical mechanisms developing antifungal resistance include mutations in the target enzyme, alterations in permeability of efflux pump proteins (ATP transport system superfamily (ABC) like CDR1p and CDR2p, and major facilitator (MFs), including MDR1 gene), and changes in modification or degradation of drugs inside the cell. Mutations in targeted enzymes and shifts in efflux pumps are shown to be of greater importance among various species of Candida (18-20). Candida albicans species can become resistant to antifungal drugs through mutations in which the gene of the target enzyme alters (21). For instance, a point mutation in ERG11 gene, which encodes the major target of azoles, 14a-demethylase, results in amino acid alteration leading to decreased enzyme affinity to azole drugs. Hence, the resistance against these agents is strengthened (22, 23). However, azole resistance can be caused by overexpression of CaERG11 as well (19, 22).
2. Objectives
This study aimed to determine antifungal susceptibility of clinical C. albicans isolates and ERG11 gene mutation. Moreover, the changes in the expression of genes responsible for azole resistance were measured using RT-PCR.
3. Methods
3.1. Patient and Fungal Isolates
Here 116 head and neck cancer patients with OPC were selected during the period of two years (Jul 2018 to Oct 2020) at Cancer Institute of Imam Khomeini Hospital in Tehran. No treatment of antifungals and prophylaxis was done on the patients, just radiotherapy. The study was informed and patient’s agreement was prepared. The data of patients, including age, sex, type of malignancy, and stage of cancer (The rate of spread of cancer in the patient's body), were obtained in patients’ sheets. Oral samples were collected with sterile swabs. Oropharyngeal candidiasis was confirmed by the existence of white plaques and certified by funding of yeasts and pseudohyphae on KOH 10% examinations and positive culture. Initial differentiation was performed by following phenotypic methods: culturing on CHROMagar Candida (CHROMagar Company, Paris, France) medium, chlamydoconidia, and germ tube production and carbohydrate assimilation. Finally, it was confirmed by multiplex PCR method (24). All Candida isolates were kept in 1.5 ml sterile 20% glycerol solution at -80°C for future use.
3.2. Antifungal Susceptibility Testing
Specific antifungal drugs containing fluconazole (Sigma-Aldrich, St. Louis, MO, USA), itraconazole (Sigma-Aldrich, St. Louis, MO, USA), voriconazole (Sigma-Aldrich, St. Louis, MO, USA), caspofungin (CFG, Merck Sharp & Dohme, Haarlem), and amphotericin B (Sigma-Aldrich, St. Louis, MO, USA) were used for this experiment according to the Clinical and Laboratory Standards Institute M27-A3 standard method (25). Various concentrations of antifungals were prepared in RPMI medium (Sigma-Aldrich, St. Louis, MO, USA) and added to the 96-well microtiter plates. Fungal suspension was prepared from 48 h cultured colonies, and inoculums were prepared in RPMI 1640 (0.5 - 2.5 ×103 cell/mL). Then, 100 µl of cell suspensions were added in each well containing 100 µl of various amounts of antifungals. The plate was kept at 37°C, and minimum inhibitory concentrations (MICs) were determined visually after 24 h (26). Candida albicans ATCC10231 was considered control. All the experiments were done in triplicate. According to the CLSI description, all isolates with a MIC of ≥ 8 µg/mL were defined as having resistance; ≥ 2 µg/mL as susceptible and 4 µg/mL as sensitive dose-dependent (SDD) to fluconazole (27).
3.3. DNA Extraction, PCR Amplification, and Sequencing
Candida albicans cell disruption was done using glass beads, and extraction of total genomic DNA was carried out using phenol-chloroform-isoamyl alcohol method (28). ERG11 was amplified by PCR using specific primers (Table 1) (29). In a reaction volume of 50 µl, amplifications were done using Taq PCR Master Mix, Ampliqon (Ampliqon, Denmark). PCRs were implemented with an initial incubation at 95°C, 5 min; 45 cycles of 95°C, 10 secs; 58°C, 150 secs; 72°C, 90 sec; followed by 72°C for 5 min (29). The ERG11 sequences were analyzed by MEGA6 software, and SNP were detected by comparing the whole ERG11 open reading frame of fluconazole susceptible strain previously submitted (XM-711668.2) by ERG11 sequence (30, 31).
The Primers Used for PCR and Real-time PCR Experiments
| Genes | Primer Sequence (5'-3') | Size (bp) |
|---|---|---|
| PCR | ||
| ERG11 | 1587 | |
| F | 5'-GTTGAAACTGTCATTGATGG-3' | |
| R | 5'-TCAGAACACTGAATCGAAAG-3' | |
| Real-time PCR | ||
| ERG11 | 91 | |
| F | 5'- AACTACTTTTGTTTATAATTTAAGATGGACTATTGA-3' | |
| R | 5'- AATGATTTCTGCTGGTTCAGTAGGT-3' | |
| CDR1 | 96 | |
| F | 5'- TTTAGCCAGAACTTTCACTCATGATT-3' | |
| R | 5'- TATTTATTTCTTCATGTTCATATGGATTGA-3' | |
| CDR2 | 80 | |
| F | 5'- GGTATTGGCTGGTCCTAATGTGA-3' | |
| R | 5'- GCTTGAATCAAATAAGTGAATGGATTAC-3' | |
| MDR1 | 83 | |
| F | 5'- TTACCTGAAACTTTTGGCAAAACA-3' | |
| R | 5'- ACTTGTGATTCTGTCGTTACCG-3' | |
| ACT1 | 85 | |
| F | 5'- TTGGTGATGAAGCCCAATCC-3' | |
| R | 5'- CATATCGTCCCAGTTGGAAACA-3' | |
3.4. Gene Expression by Real-time PCR
The ERG11, CDR1, CDR2, and MDR1 gene expressions were evaluated in 42 fluconazole-resistant, susceptible-dose-dependent (SDD), and susceptible C. albicans isolates from OPC head and neck cancer patients as follows. Whole RNA extraction was done from the homogenized fungal cells by GITC (Guanidinium Isothiocyanate) reagent and glass beads, then treatment with RNase-free DNase was done (Thermo Fisher Scientific, USA) (32). From a total of 1,000 ng RNA, single-stranded cDNA was prepared using Revert Aid M-MuLV and random hexamer primers in cDNA synthesis kit (Yekta Tajhiz, Iran). Real-time PCR was performed (33) by SYBR green master mix (Sina Clone, Iran). The final volume of each reaction was 25 µL performed by a Rotor gene 6,000 (Corbett System). The specific primer sets were showed in Table 1 (34). Real-time PCR was performed by the following program: 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min (35). All tests were done in triplicate. The results were determined by relative quantification, using ACT1 expression as the reference gene. Gene folding change levels were measured by 2−ΔCT method. Determination of fold increases (FI) was done by the relative threshold method (2−ΔΔCT) (33).
3.5. Statistical Analysis
The data of gene expression were considered for the analysis of variance (One-way ANOVA) in Tukey range. The differences with a P-value < 0.05 were considered significant. For P-value evaluation, one-way ANOVA test using GraphPad Prism 6 (San Diego, CA, USA) was used.
4. Results
4.1. Clinical Data
In this study, 217 Candida isolates were obtained from 116 OPC patients with head and neck cancer in Imam Khomeini Hospital, institute of cancer in Tehran, for about two years (Jul 2018 to Oct 2020). Of the total isolates, 125 isolates were C. albicans, and 92 isolates were Candida non-albicans. Only C. albicans isolates were included in this study. Mixed isolates were observed in 5 patients. The fungal yield of each sample was 97 - 15 ± 5 CFU/plate after culturing on Sabouraud dextrose agar (SDA) medium. The results of initial differentiation showed that seven (5.6%) and nine (7.2%) C. albicans isolates did not produce any germ tube and chlamydoconidium, respectively. Also, four (3.2%) C. albicans isolates represented white color on CHROMagar Candida medium. All of these isolates were detected as C. albicans by multiplex PCR (the total number of C. albicans isolates was 125 isolates). Our data showed that 73 (58.4%) C. albicans isolates were obtained from males, and 52 (41.6%) C. albicans isolates were obtained from females. More patients were in stage 2 (57.6%), followed by stage 1 (32%) and stage 3 (10.4%). The patients’ ages ranged from 12 to 93 years, and the average age was 52. Most of the patients had face basal cell carcinoma (BCC, 45%), maxillary squamous cell carcinoma (SCC, 28%), tongue SCC (19%), and the rest of the patients (8%) had other head and neck malignancies.
4.2. Antifungal Susceptibility Profiles
The results of antifungal susceptibility of C. albicans isolates showed that 15 isolates (12%) from 125 C. albicans isolates represented a reduction in susceptibility to fluconazole. Moreover, 12 (9.6%) isolates were considered to be SDD, and 98 (78.4%) isolates were susceptible to fluconazole. Moreover, 14 isolates (11.2%) displayed susceptibility reduction to voriconazole, and 16 isolates (12.8%) represented reduced susceptibility to itraconazole. In addition, reduced sensitivity to amphotericin B was observed in 23 (18.4%) isolates and in relation to caspofungin in 13 (10.4%) isolates. The demographic data and details of all C. albicans isolates with antifungal susceptibility are shown in Tables 2 and 3.
Antifungal Susceptibility Results of 125 Clinical Candida albicans Isolates Recovered from Oropharyngeal Candidiasis in Patients Suffering from Head and Neck Cancer a
| Antifungal Agents | Category, No. (%) | CBPs (µg/mL) | ECV (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| S/WT | SDD/I | R/NWT | Gmean (µg/mL) | S | SDD | I | R | ||
| Fluconazole | 98 (78.4) | 12 (9.6) | 15 (12) | 4.018 | ≤ 2 | 4 | - | ≥ 8 | - |
| Itraconazole | 68 (54.4) | 41 (32.8) | 16 (12.8) | 0.371 | ≥ 0.12 | 0.25 - 0.5 | - | ≥ 1 | - |
| Voriconazole | 74 (59.2) | 37 (29.6) | 14 (11.2) | 0.314 | ≥ 0.12 | - | 0.25 - 0.5 | ≥ 1 | - |
| Caspofungin | 87 (69.6) | 25 (20.0) | 13 (10.4) | 0.373 | ≥ 0.25 | - | 0.5 | ≥ 1 | - |
| Amphotericin B | 102 (81.6) | - | 23 (18.4) | 0.739 | - | - | - | - | 2 |
In vitro Azole Susceptibility, ERG11 Sequence, and Gene Expression Levels for the Clinical Isolates of Candida albicansa
| No. | Isolate | Accession No. | MIC (µg/mL) | Amino Acid Substitution(s) in ERG11 | Gene Overexpression | ||||
|---|---|---|---|---|---|---|---|---|---|
| FLZ | VRC | ITZ | AMB | CAS | |||||
| 1 | 2pr-55 | OM774349 | > 64 | 4.00 | 8.00 | 4.00 | 2.00 | D116E, K128T, G464S | CDR1, CDR2, ERG11 |
| 2 | 2pr-22 | OM774363 | > 64 | 0.50 | 0.50 | 0.50 | 0.125 | A114S, Y257H, K344E, R523G | MDR1 |
| 3 | 2pr-73 | OM774368 | > 64 | 0.125 | 0.50 | 0.50 | 1.00 | D116E, E266D, G464S | CDR1, CDR2 |
| 4 | 2pr-125 | OM774374 | > 64 | 1.00 | 2.00 | 0.50 | 0.125 | A114S, K128T, K344E, R522G | CDR1, CDR2, MDR1 |
| 5 | 2pr-114 | OM774373 | > 64 | 0.50 | 1.00 | 0.50 | 0.50 | D116E, K128T, E266D, G464S | CDR1, CDR2 |
| 6 | 2pr-142 | OM774378 | 64 | 0.50 | 0.125 | 1:00 | 0.0312 | D116E, K128T E266D, | CDR2 |
| 7 | 2pr-145 | OM774379 | 32 | 0.50 | 0.25 | 2.00 | 0.25 | A114S, E266D | CDR1, ERG11 |
| 8 | 2pr-195 | OM774384 | 8.00 | 1.00 | 0.125 | 0.0312 | 0.50 | V488I, W520C | CDR1, CDR2 |
| 9 | 2pr-354 | OM774390 | 8.00 | 0.50 | 0.125 | 1.00 | 1.00 | E266D, V488I, W520C | CDR1, CDR2 |
| 10 | 2pr-9 | OM774357 | 8.00 | 1.00 | 0.125 | 4.00 | 0.25 | D116E, E266D, P375A | CDR2 |
| 11 | 2pr-135 | OM774376 | 8.00 | 0.0625 | 0.125 | 1:00 | 0.25 | D116E, E266D, V488I | CDR2 |
| 12 | 2pr-15 | OM774360 | 8.00 | 0.50 | 1.00 | 0.125 | 1.00 | E266D, V488I, W520C | CDR2 |
| 13 | 2pr-56 | OM774361 | 8.00 | 0.125 | 0.50 | 0.50 | 0.125 | G59S, D116E, E266D, | CDR1 |
| 14 | 2pr-42 | OM774364 | 8:00 | 0.50 | 1.00 | 0.125 | 0.125 | E266D, W520C | CDR1 |
| 15 | 2pr-278 | OM774386 | 8.00 | 0.50 | 0.50 | 2.00 | 0.125 | G59S, E266D | CDR1, CDR2 |
| 16 | 2pr-58 | OM774362 | 4.00 | 0.125 | 0.125 | 1.00 | 0.0312 | V51L | CDR1 |
| 17 | 2pr-65 | OM774354 | 4.00 | 0.50 | 0.50 | 1.00 | 0.25 | D116E, E266D | CDR1 |
| 18 | 2pr-47 | OM774366 | 4.00 | 1.00 | 2.00 | 0.50 | 0.50 | D116E, E266D | CDR1, CDR2 |
| 19 | 2pr-87 | OM774367 | 4.00 | 1.00 | 0.50 | 0.125 | 0.25 | D116E, E266D | CDR2 |
| 20 | 2pr-77 | OM774369 | 4:00 | 0.50 | 2.00 | 0.125 | 1.00 | D116E, E266D | CDR2 |
| 21 | 2pr-57 | OM774350 | 4.00 | 0.25 | 0.50 | 2.00 | 0.50 | D116E, E266D, | CDR1, |
| 22 | 2pr-59 | OM774351 | 4.00 | 0.0312 | 2.00 | 0.0625 | 8.00 | E266D | CDR1, CDR2 |
| 23 | 2pr-61 | OM774352 | 4.00 | 0.125 | 0.125 | 0.50 | 0.25 | D116E, E266D | CDR2 |
| 24 | 2pr-63 | OM774353 | 4.00 | 0.50 | 2.00 | 0.125 | 0.25 | D116E, E266D | CDR1, CDR2 |
| 25 | 2pr-7 | OM774356 | 4.00 | 0.25 | 0.125 | 0.50 | 0.25 | E266D | CDR2 |
| 26 | 2pr-36 | OM774365 | 4.00 | 1.00 | 0.50 | 1.00 | 0.25 | D116E, E266D | CDR1, CDR2, |
| 27 | 2pr-83 | OM774371 | 4.00 | 0.50 | 0.125 | 0.125 | 0.0625 | D116E, E266D | CDR2, |
| 28 | 2pr-329 | OM774387 | 0.0312 | 0.125 | 0.50 | 1.00 | 0.125 | D116E, E266D | - |
| 29 | 2pr-332 | OM774388 | 0.125 | 0.50 | 0.125 | 0.0312 | 0.50 | D116E, E266D | - |
| 30 | 2pr-339 | OM774389 | 1.00 | 0.125 | 0.125 | 0.50 | 0.25 | D116E | - |
| 31 | 2pr-11 | OM774358 | 2.00 | 0.50 | 0.50 | 1.00 | 1.00 | E266D, D504A | - |
| 32 | 2pr-13 | OM774359 | 0.50 | 0.125 | 0.50 | 1.00 | 0.50 | D116E | - |
| 33 | 2pr-108 | OM774372 | 1.00 | 0.50 | 0.50 | 1.00 | 2.00 | E266D | - |
| 34 | 2pr-149 | OM774381 | 2.00 | 0.125 | 0.50 | 0.125 | 0.0312 | D116E, E266D | - |
| 35 | 2pr-168 | OM774383 | 2.00 | 0.0312 | 0.50 | 0.0625 | 0.25 | D116E | - |
| 36 | 2pr-201 | OM774385 | 1.00 | 0.50 | 0.125 | 1:00 | 0.125 | E266D | - |
| 37 | 2pr-129 | OM774375 | 0.125 | 0.0312 | 0.125 | 1.00 | 0.25 | - | - |
| 38 | 2pr-93 | OM774370 | 0.0312 | 1.00 | 0.125 | 1:00 | 0.125 | - | - |
| 39 | 2pr-136 | OM774377 | 0.50 | 0.50 | 0.50 | 0.125 | 0.0312 | - | - |
| 40 | 2pr-163 | OM774382 | 0.50 | 0.0312 | 0.50 | 1.00 | 0.25 | - | - |
| 41 | 2pr-67 | OM774355 | < 0.0312 | 1.00 | 0.125 | < 0.0312 | > 16 | - | - |
| 42 | 2pr-146 | OM774380 | 2.00 | 0.125 | 0.0312 | 0.125 | 0.25 | - | - |
4.3. ERG11 Sequencing Results
The ERG11 gene sequence of 42 C. albicans isolates, 15 fluconazole-resistant, 12 SDD, and 15 susceptible isolates were done and deposited in GenBank database under accession number OM774349-OM774390. ERG11 coding region was amplified by PCR in the size of 1,640 bp. The results of ERG11 sequencing from 42 C. albicans isolates are shown in Table 3. We found 34 mutations that 20 of which were silent mutations and did not change amino acid. Fourteen missense mutations were detected in 42 R/SDD/S C. albicans isolates. Among the 14 missense mutations identified, nine mutations had been identified previously, including D116E, K128T, G465S, E266D, V488I, A114S, Y257H, K344E, and R523G. Five mutations were reported newly, including D504A, P375A, W520C, G59S, and V51L (Table 3). D116E and E266D are the most common mutations among 42 C. albicans isolates.
4.4. Efflux Transporters Gene Expression Levels
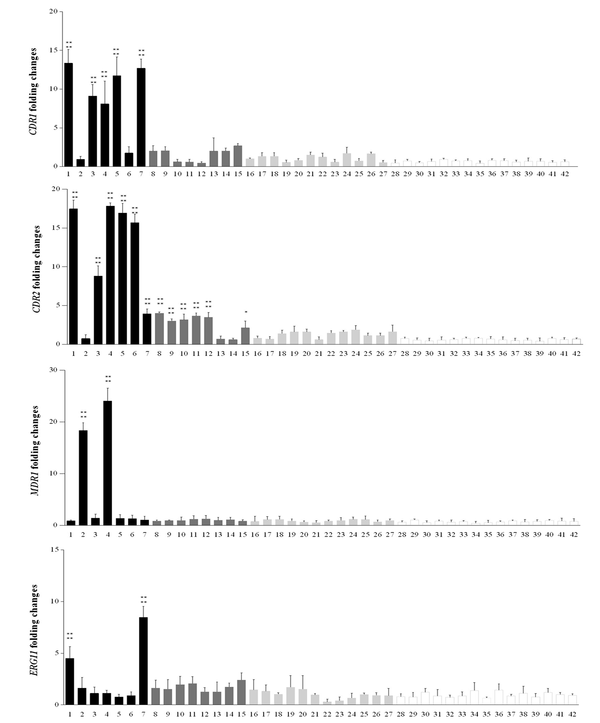
The expression levels of CDR1, CDR2, ERG11, and MDR1 for 42 clinical Candida isolates were measured. The expression of the genes in C. albicans strains was compared with the mean expression level of 15 fluconazole-susceptible isolates by quantitative PCR. At least 2-fold increased overexpression was observed (36). The results showed that five (33.3%) resistant isolates with MIC ≥ 32 µg/mL represented increased expression levels of CDR1 and CDR2, five isolates with MIC 8 µg/mL showed CDR1 and six isolates represented CDR2 expression levels upper than 2-fold of the mean of susceptible isolates. MDR1 and ERG11 expression levels were increased in two (13.3 %) resistant isolates with MIC ≥ 32 µg/mL (Table 3, Figure 1). Statistical analysis represented a significant difference in the mean of gene expression of resistance isolates in comparison to the mean of gene expression in susceptible isolates, P < 0.0001 (Figure 1).
CDR1, CDR2, MDR1, and ERG11 fold expression levels in azole-resistance with MIC ≥ 32 µg/mL (n = 7; black bars) and MIC = 8 µg/mL (n = 8 dark grey bars), SDD with MIC = 4 µg/mL (n = 12; light grey bars) and susceptible with MIC < 2 (n = 15; white bars) groups of Candida albicans. Each of the target gene expression levels was measured by the 2-ΔΔCt method using ACT1 housekeeping gene as the internal control. The process of each sample was done in triplicate. Error bars show the standard deviations; *Statistically significant difference with the mean of gene expression in susceptible isolates, P < 0.0001.
5. Discussion
Oropharyngeal candidiasis is considered to be the most frequent oral clinical Candida spp. manifestation in people with head and neck malignancies (37). For oropharyngeal candidiasis treatment, Azoles have been the choice (34). The high frequency of azole resistance and the speed at which C. albicans resistance is acquired is a crucial concern for clinicians, especially in the case of immunocompromised patients (38). Azole resistance in C. albicans might be caused by overexpression of genes, which encode efflux pumps or might result from mutations in or overexpression of ERG11 (34). Since there are a few studies about antifungal resistance of species of C. albicans that are relevant to OPC in Iranian patients with head and neck cancer, the current study was conducted to evaluate antifungal susceptibility patterns and molecular mechanisms of these isolates in Institute of Cancer in Imam Khomeini Hospital located in Tehran. Among 125 C. albicans isolates, 27 isolates were R, and SDD to fluconazole, and 98 isolates were susceptible. Therefore overall, 42 isolates (27 R and SDD beside 15 susceptible isolates) were considered to be done for ERG11 sequence and real-time PCR analysis.
Our results illustrated 14 missense mutations in ERG11 gene that substituted amino acid sequence. Among them, D116E and E266D were expressed in all of the resistant isolates, including SDD and some susceptible C. albicans isolates, which is consistent with other studies, showing that these mutations probably have no effects in reduction of azole susceptibility (31, 39, 40). It has been reported (30) E266D substitution in resistant isolates of C. albicans, while other studies demonstrated this amino acid substitution in both azole resistance and azole susceptible isolates. It has been demonstrated that the amino acid substitution D116E was not associated with the azole-resistant phenotype (39). Furthermore, previous studies showed that A114S, Y257H, K128T, and V488I mutations were responsible for fluconazole susceptibility reduction in C. albicans isolates (38, 41-43).
In our study, C. albicans resistant isolates represented A114S, Y257H, and K128T amino acid substitutions (K128T substitution was found in 14 isolates, A114S was found in four isolates and Y257H was found in one isolate of C. albicans), which strongly suggests that these are associated with the azole-resistant phenotype. Combined substitutions of Y257H and A114S have been informed in fluconazole-resistant isolates. These substitutions were demonstrated to increase fluconazole resistance as well. Since the location of A114S is next to the ERG11p substrate channel, the interference with active site binding or inhibitor may occur due to its mutations; however, Y257H does not appear to affect the ERG11p for azoles since it is located in the G helix, which is far away from substrate channel of the protein. Therefore, further verification of Y257H mutation association with azole resistance must be carried out (30). Here we found new amino acid substitutions, including D504A, P375A, W520C, G59S, and V51L. Interestingly, D504A was observed in susceptible isolates, but others were found in R and SDD isolates (MIC ≥ 8 µg/mL and MIC = 4 µg/mL), suggesting that they contributed to reduced susceptibility isolates.
Real-time PCR was carried out to discover the expression levels of CDR1, CDR2, ERG11, and MDR1 genes for all resistant, SDD, and susceptible isolates. Our results indicated that CDR2 gene showed increased expression in more resistant isolates compared with other tested genes, followed by the CDR1 gene. Studies have demonstrated that expression of CDR1 and CDR2 were elevated in the azole-resistant isolates, in comparison with isolates susceptible to azole, since CDR2 expression was at higher levels compared to CDR1 (38). Interestingly, our results showed that the expression level of CDR2 gene was higher than CDR1.
In this study, gene expression increased by two folds compared with the mean of susceptible isolates that was considered the target gene overexpression. Five isolates with MIC ≥ 32 represented overexpression in CDR1 with the range of 13.36 to 8.09, and the expression level of CDR1 in five isolates with MIC = 8 µg/mL was 2 to 2.7. Moreover, in the eight SDD isolates (MIC = 4 µg/mL), the expression level of CDR1 was 1 to 1.6 (The mean expression level in susceptible isolates was 0.666) (Figure 1). Also, five C. albicans isolates showed overexpression range of 17.8 to 8.8 in CDR2 gene (The mean CDR2 expression level was 0.615 in susceptible isolates). In some isolates with MIC = 8 µg/mL and 4 µg/mL, the expression levels more than twice as high as the average of susceptible isolates were seen as the expression level was between three to four in six isolates with MIC = 8 µg/mL (Table 3, Figure 1).
The overexpression of ERG11, encoding lanosterol demethylase, a key enzyme in the ergosterol biosynthesis pathway, is a significant reason for fluconazole resistance in C. albicans. Flowers et al. demonstrated that ERG11 overexpression was observed in most of the fluconazole-resistant isolates. They suggested that ERG11 overexpression is a common contributor to resistance in C. albicans (36). Furthermore, Liu et al. reported that ERG11 was not overexpressed in fluconazole-resistant C. albicans isolates. They suggested that ERG11 overexpression is not crucial for azole resistance induction in C. albicans (38). However, here we demonstrated that ERG11 overexpression was observed just in two fluconazole-resistant isolates (expression level ranges from 8 to 4.5) but not in all of them. It has been demonstrated that resistance to fluconazole results from excessive MDR1 expression.
It shed light on a principal mechanism of clinical resistant isolates (44). Our study also indicated that the expression of MDR1 increased more than ERG11 in isolates that showed reduced sensitivity to fluconazole (Table 3, Figure 1). It has been shown that fluconazole-resistant isolates represented high levels of gene expression in MDR1 (44). MDR1 gene overexpression was seen in two resistant isolates in the range of 24.06 to 18.34. This work has some limitations, such as our inability to analyze the molecular epidemiology of C. albicans isolates to determine the relationship between fluconazole resistance and genetic affinity of the clinical C. albicans isolates.
Taken together, as there is little information about antifungal resistance pattern of C. albicans clinical isolates from OPC in Iranian head and neck cancer patients, our study aimed to evaluate fluconazole resistance mechanism of these clinical isolates and to find a correlation between sex and age of these patients and the drug resistance for the first time. Moreover, in this study, there was no relationship between drug resistance, cancer type, and sex and age of patients, suggesting that a large sample size might be needed to find the relationship between them.
5.1. Conclusions
This study demonstrated that mutation in ERG11 gene was the most causative mechanism for fluconazole resistance in C. albicans isolates that were obtained from patients with head and neck cancer suffering from oropharyngeal candidiasis. Additionally, we found that caspofungin was the effective antifungal substance in fluconazole resistance situations for C. albicans infection in these isolates. Identification of drug resistance mechanisms and antifungal susceptibility patterns are considered helpful in using appropriate antifungal drugs and preventing antifungal resistance.
Acknowledgements
References
-
1.
Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20(1):133-63. [PubMed ID: 17223626]. [PubMed Central ID: PMC1797637]. https://doi.org/10.1128/CMR.00029-06.
-
2.
Ganguly S, Mitchell AP. Mucosal biofilms of Candida albicans. Curr Opin Microbiol. 2011;14(4):380-5. [PubMed ID: 21741878]. [PubMed Central ID: PMC3159763]. https://doi.org/10.1016/j.mib.2011.06.001.
-
3.
Calderone RA. Candida and candidiasis. Emerg Infect Dis. 2002;8(8):66-75. [PubMed ID: 2732517]. [PubMed Central ID: PMC2732517]. https://doi.org/10.3201/eid0808.020059.
-
4.
Pappas PG, Rex JH, Sobel JD, Filler SG, Dismukes WE, Walsh TJ, et al. Guidelines for treatment of candidiasis. Clin Infect Dis. 2004;38(2):161-89. [PubMed ID: 14699449]. https://doi.org/10.1086/380796.
-
5.
Wenzel RP. Nosocomial candidemia: risk factors and attributable mortality. Clin Infect Dis. 1995;20(6):1531-4. [PubMed ID: 7548504]. https://doi.org/10.1093/clinids/20.6.1531.
-
6.
Fox EP, Nobile CJ. The role of Candida albicans biofilms in human disease. In: Dietrich LA, Friedmann TS, editors. Candida albicans symptoms, causes and treatment options. Nova Science Publishers; 2013. p. 1-24.
-
7.
Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11(1):30-6. [PubMed ID: 12526852]. https://doi.org/10.1016/s0966-842x(02)00002-1.
-
8.
Talapko J, Juzbasic M, Matijevic T, Pustijanac E, Bekic S, Kotris I, et al. Candida albicans-the virulence factors and clinical manifestations of infection. J Fungi (Basel). 2021;7(2). [PubMed ID: 33499276]. [PubMed Central ID: PMC7912069]. https://doi.org/10.3390/jof7020079.
-
9.
Silverman SJ, Luangjarmekorn L, Greenspan D. Occurrence of oral Candida in irradiated head and neck cancer patients. J Oral Med. 1984;39(4):194-6. [PubMed ID: 6389802].
-
10.
Chen TY, Webster JH. Oral monilia study on patients with head and neck cancer during radiotherapy. Cancer. 1974;34(2):246-9. [PubMed ID: 4604859]. https://doi.org/10.1002/1097-0142(197408)34:2<246::aid-cncr2820340203>3.0.co;2-j.
-
11.
Laudenbach JM, Epstein JB. Treatment strategies for oropharyngeal candidiasis. Expert Opin Pharmacother. 2009;10(9):1413-21. [PubMed ID: 19505211]. https://doi.org/10.1517/14656560902952854.
-
12.
Gafter-Gvili A, Vidal L, Goldberg E, Leibovici L, Paul M. Treatment of invasive candidal infections: systematic review and meta-analysis. Mayo Clin Proc. 2008;83(9):1011-21. [PubMed ID: 18775201]. https://doi.org/10.4065/83.9.1011.
-
13.
Katragkou A, Roilides E, Walsh TJ. Role of echinocandins in fungal biofilm-related disease: vascular catheter-related infections, immunomodulation, and mucosal surfaces. Clin Infect Dis. 2015;61 Suppl 6:S622-9. [PubMed ID: 26567280]. https://doi.org/10.1093/cid/civ746.
-
14.
Rex JH, Walsh TJ, Sobel JD, Filler SG, Pappas PG, Dismukes WE, et al. Practice guidelines for the treatment of candidiasis. Infectious Diseases Society of America. Clin Infect Dis. 2000;30(4):662-78. [PubMed ID: 10770728]. https://doi.org/10.1086/313749.
-
15.
Goa KL, Barradell LB. Fluconazole. An update of its pharmacodynamic and pharmacokinetic properties and therapeutic use in major superficial and systemic mycoses in immunocompromised patients. Drugs. 1995;50(4):658-90. [PubMed ID: 8536553]. https://doi.org/10.2165/00003495-199550040-00007.
-
16.
Grant SM, Clissold SP. Fluconazole. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in superficial and systemic mycoses. Drugs. 1990;39(6):877-916. [PubMed ID: 2196167]. https://doi.org/10.2165/00003495-199039060-00006.
-
17.
Alexander BD, Perfect JR. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs. 1997;54(5):657-78. [PubMed ID: 9360056]. https://doi.org/10.2165/00003495-199754050-00002.
-
18.
White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11(2):382-402. [PubMed ID: 9564569]. [PubMed Central ID: PMC106838]. https://doi.org/10.1128/CMR.11.2.382.
-
19.
Chen LM, Xu YH, Zhou CL, Zhao J, Li CY, Wang R. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Med Res. 2010;38(2):536-45. [PubMed ID: 20515567]. https://doi.org/10.1177/147323001003800216.
-
20.
Marger MD, Saier MJ. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18(1):13-20. [PubMed ID: 8438231]. https://doi.org/10.1016/0968-0004(93)90081-w.
-
21.
Prasad R, Nair R, Banerjee A. Emerging mechanisms of drug resistance in Candida albicans. Prog Mol Subcell Biol. 2019;58:135-53. [PubMed ID: 30911892]. https://doi.org/10.1007/978-3-030-13035-0_6.
-
22.
Spettel K, Barousch W, Makristathis A, Zeller I, Nehr M, Selitsch B, et al. Analysis of antifungal resistance genes in Candida albicans and Candida glabrata using next generation sequencing. PLoS One. 2019;14(1). e0210397. [PubMed ID: 30629653]. [PubMed Central ID: PMC6328131]. https://doi.org/10.1371/journal.pone.0210397.
-
23.
Ying Y, Zhao Y, Hu X, Cai Z, Liu X, Jin G, et al. In vitro fluconazole susceptibility of 1,903 clinical isolates of Candida albicans and the identification of ERG11 mutations. Microb Drug Resist. 2013;19(4):266-73. [PubMed ID: 23484590]. https://doi.org/10.1089/mdr.2012.0204.
-
24.
Arastehfar A, Fang W, Pan W, Lackner M, Liao W, Badiee P, et al. YEAST PANEL multiplex PCR for identification of clinically important yeast species: stepwise diagnostic strategy, useful for developing countries. Diagn Microbiol Infect Dis. 2019;93(2):112-9. [PubMed ID: 30377018]. https://doi.org/10.1016/j.diagmicrobio.2018.09.007.
-
25.
Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. CLSI document M27-A3 and Supplement S. Wayne, USA: Clinical and Laboratory Standards Institute; 2008.
-
26.
Jahn B, Martin E, Stueben A, Bhakdi S. Susceptibility testing of Candida albicans and Aspergillus species by a simple microtiter menadione-augmented 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay. J Clin Microbiol. 1995;33(3):661-7. [PubMed ID: 7751374]. [PubMed Central ID: PMC228010]. https://doi.org/10.1128/jcm.33.3.661-667.1995.
-
27.
Pfaller MA, Diekema DJ. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50(9):2846-56. [PubMed ID: 22740712]. [PubMed Central ID: PMC3421803]. https://doi.org/10.1128/JCM.00937-12.
-
28.
Yamada Y, Makimura K, Merhendi H, Ueda K, Nishiyama Y, Yamaguchi H, et al. Comparison of different methods for extraction of mitochondrial DNA from human pathogenic yeasts. Jpn J Infect Dis. 2002;55(4):122-5. [PubMed ID: 12403909].
-
29.
Xisto MI, Caramalho RD, Rocha DA, Ferreira-Pereira A, Sartori B, Barreto-Bergter E, et al. Pan-azole-resistant Candida tropicalis carrying homozygous erg11 mutations at position K143R: a new emerging superbug? J Antimicrob Chemother. 2017;72(4):988-92. [PubMed ID: 28065893]. https://doi.org/10.1093/jac/dkw558.
-
30.
Wang B, Huang LH, Zhao JX, Wei M, Fang H; Wang, et al. ERG11 mutations associated with azole resistance in Candida albicans isolates from vulvovaginal candidosis patients. Asian Pac J Trop Biomed. 2015;5(11):909-14. [PubMed ID: 23480635]. https://doi.org/10.1016/j.apjtb.2015.08.002.
-
31.
Lai MH, Kirsch DR. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17(2):477–498. [PubMed ID: 2644625]. [PubMed Central ID: PMC331629]. https://doi.org/10.1093/nar/17.2.804.
-
32.
Rezaie S, Ban J, Mildner M, Poitschek C, Brna C, Tschachler E. Characterization of a cDNA clone, encoding a 70 kDa heat shock protein from the dermatophyte pathogen Trichophyton rubrum. Gene. 2000;241(1):27-33. [PubMed ID: 10607895]. https://doi.org/10.1016/s0378-1119(99)00475-8.
-
33.
Losberger C, Ernst JF. Sequence of the Candida albicans gene encoding actin. Nucleic Acids Res. 1989;17(22):9488. [PubMed ID: 2685762]. [PubMed Central ID: PMC335165]. https://doi.org/10.1093/nar/17.22.9488.
-
34.
Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother. 2004;48(6):2124-31. [PubMed ID: 15155210]. [PubMed Central ID: PMC415610]. https://doi.org/10.1128/AAC.48.6.2124-2131.2004.
-
35.
Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, et al. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot Cell. 2007;6(11):2122-38. [PubMed ID: 17905926]. [PubMed Central ID: PMC2168409]. https://doi.org/10.1128/EC.00327-07.
-
36.
Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, et al. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell. 2012;11(10):1289-99. [PubMed ID: 22923048]. [PubMed Central ID: PMC3485914]. https://doi.org/10.1128/EC.00215-12.
-
37.
Suryawanshi H, Ganvir SM, Hazarey VK, Wanjare VS. Oropharyngeal candidosis relative frequency in radiotherapy patient for head and neck cancer. J Oral Maxillofac Pathol. 2012;16(1):31-7. [PubMed ID: 22438640]. [PubMed Central ID: PMC3303519]. https://doi.org/10.4103/0973-029X.92970.
-
38.
Liu JY, Shi C, Wang Y, Li WJ, Zhao Y, Xiang MJ. Mechanisms of azole resistance in Candida albicans clinical isolates from Shanghai, China. Res Microbiol. 2015;166(3):153-61. [PubMed ID: 25748216]. https://doi.org/10.1016/j.resmic.2015.02.009.
-
39.
Xiang MJ, Liu JY, Ni PH, Wang S, Shi C, Wei B, et al. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013;13(4):386-93. [PubMed ID: 23480635]. https://doi.org/10.1111/1567-1364.12042.
-
40.
Wang H, Kong F, Sorrell TC, Wang B, McNicholas P, Pantarat N, et al. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiol. 2009;9:167. [PubMed ID: 19682357]. [PubMed Central ID: PMC2782262]. https://doi.org/10.1186/1471-2180-9-167.
-
41.
Teo JQ, Lee SJ, Tan AL, Lim RS, Cai Y, Lim TP, et al. Molecular mechanisms of azole resistance in Candida bloodstream isolates. BMC Infect Dis. 2019;19(1):63. [PubMed ID: 30654757]. [PubMed Central ID: PMC6337757]. https://doi.org/10.1186/s12879-019-3672-5.
-
42.
Berkow EL, Lockhart SR. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist. 2017;10:237-45. [PubMed ID: 28814889]. [PubMed Central ID: PMC5546770]. https://doi.org/10.2147/IDR.S118892.
-
43.
Manastir L, Ergon MC, Yucesoy M. Investigation of mutations in Erg11 gene of fluconazole resistant Candida albicans isolates from Turkish hospitals. Mycoses. 2011;54(2):99-104. [PubMed ID: 19732347]. https://doi.org/10.1111/j.1439-0507.2009.01766.x.
-
44.
Dunkel N, Liu TT, Barker KS, Homayouni R, Morschhauser J, Rogers PD. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot Cell. 2008;7(7):1180-90. [PubMed ID: 18487346]. [PubMed Central ID: PMC2446669]. https://doi.org/10.1128/EC.00103-08.