1. Background
Exointestinal pathogenic Escherichia coli (EPEC) strains can cause urinary tract infections, bacteremia, or neonate meningitides (1). Exointestinal pathogenic strains harbor factors that are important in effective attachment. These bacteria are the primary cause of community-acquired urinary tract infections (UTI) (70% - 95%) and a large portion of nosocomial UTIs (50%), accounting for substantial medical costs and morbidity worldwide (2). Uropathogenic Escherichia coli (UPEC) strains take an advantage of the host behavior and can employ a diverse repertoire of virulence factors to colonize the urinary tract (3). In some cases, UPEC isolates may be disseminated by contaminated food or sexually activity. Due to variation in virulence genotypes of uropathogenic strains of E. coli based on geographical regions (4), conducting surveys on isolates characteristics in every region is of great importance.
Biofilm is a bacterial community with structural and physiological changes compared to planktonic bacteria, which have novel properties. These changes are related to enhancing bacterial surveillance because of disinfectant and antibiotic resistance. Identifying key biofilm determinants in several bacteria is required to achieve preventative strategies in initial bacterial adhesion, maturation of biofilm, or enhancement of antibiotic susceptibility of bacterial community in biofilm (5, 6). In the differentiated superficial umbrella cells that line the lumen of the bladder, UPEC are able to break into the host cell cytosol and rapidly multiply, forming large intracellular biofilm-like communities that can contain several thousand bacteria.
Biofilm development could be stopped by removing any of the followings: microbes, slime- exopolysaccharides, and surface (7). Different superficial appendices of organisms are related to the first stage of biofilm formation and attachment to eukaryotic host cells such as flagella, fimbriae, autotransporter proteins, curli, F conjugative pilus, and exopolysaccharide production (8). At the first stage of biofilm formation, superficial appendages such as flagella are synthesized to lose attachment of bacteria, which is a determinant of biofilm architecture. Then, for tight attachment, which is important in biofilm formation, the synthesis of flagella suppresses, and various organelles such as curli fimbriae and Type I fimbriae, encoded by csgA and FimA genes, increase (9, 10). Curli is a thin, wiry long protein fiber on a surface of some cells (11). An increased ability to bind to abiotic surfaces such as coverslips, glass, and polystyrene or biotic surface (such as intestinal cells) by some curli-producing E. coli was reported compared to non-curli-producing strains (12). Several studies have reported that the association of some attachment factor expressions such as curli biofilm formation by extraintestinal E. coli can be associated with the biofilm production (13). Biofilm is formed by numerous species of microorganisms and favored under different environmental conditions.
2. Objectives
Because of the important role of biofilm in chronic UTI, in this study, we aimed at focusing on human uropathogenic E. coli biofilm formation by microtiter plate crystal violet and its relation to csgA and FimA genes.
3. Methods
3.1. Isolation of Pathogenic Escherichia coli
The studied E. coli in this research were isolated from the urinary infections cases of humans in Ahvaz oil big hospital. For this purpose, collected samples were cultured in blood agar (Merck, Germany) and incubated for 24 hours in 37 centigrade degree. Then, suspicious colonies were collected and streaked on blood agar again and detected by catalase, oxidase, gram stain, and biochemical tests including Triple Sugar Iron Agar (Merck, Germany), MacConkey (Merck, Germany), Sulfur-Indole-Motility (Biolab, India), Ureas (liofilchem, USA), Simmon Citrate (HiMedia, India), Lysine Iron Agar (Merck, Germany), Phenylalanin Deaminase (Merck, Germany), and Methyl Red-Vogues Prosquer (HiMedia, India).
3.2. Biofilm Formation Assay
The isolates biofilm production ability was assessed by modified technique of polystyrene microtiter plate, which was described by Stepanovic et al. Escherichia coli (ATCC 25922) standard was used as biofilm positive control. The isolates were grown on brain heart infusion (BHI) broth, supplemented with 3% sucrose (Merck, Germany), and incubated at 37°C for 24 hours. Then, 100 μL of overnight culture was transferred to 900 μL of fresh BHI broth, with 3% sucrose until observing MacFarland scale of 0.5. Then, in triplicate, 200 microliter of each isolate suspension was put in each well of microplate (Maxwell, China) and incubated at 37°C for 24 hours. Non-inoculated supplemented BHI was used as negative control in triplicate. After incubation of the plate for 24 hours in 37°C, the bacterial suspension was aspirated and washed by sterile physiological saline 3 times. After drying the plate at room temperature, 200 μL of methanol per well was added for fixation. After 15 minutes, the plate was aspirated and dried at room temperature, then, it was stained by 200 μL of 2% solution of Hucker’s crystal violet. After 5 minutes, the plate was washed by distilled water and dried at room temperature. After addition of 200 μL of discoloring solution (ethanol- aceton) for 15 minutes, absorbance was read using an ELISA plate reader (Biotek SX2, USA) at 600 nanometer.
The optical density (ODs) of each strain was obtained by the arithmetic mean of the absorbance of 3 wells, and this value was compared with the mean absorbance of negative controls (ODnc). The following classification was used to determine biofilm formation: no biofilm production (ODs < ODnc), weak biofilm production (ODnc < ODs < 2.ODnc), moderate biofilm production (2.ODnc < ODs < 4.ODnc), and strong biofilm production (4.ODnc < ODs) (14, 15).
3.3. Multiplex PCR Assay
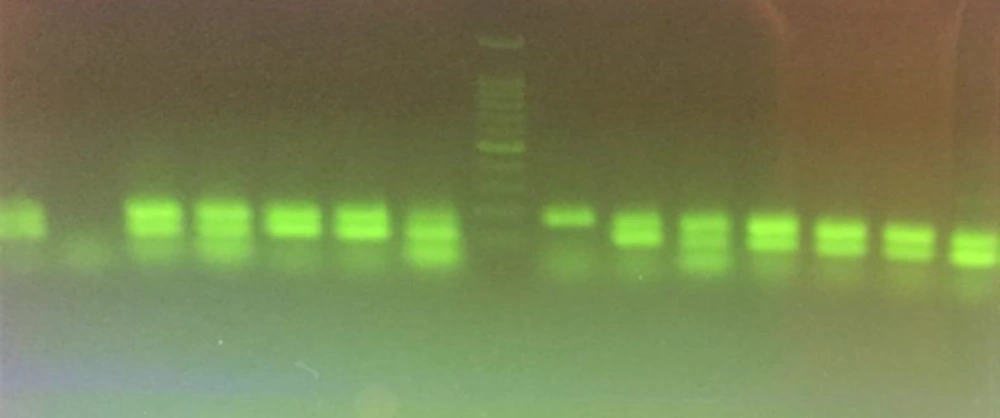
To identify the studied genes (csgA and fimA) in the isolated E. coli, PCR assays were performed according to Oliveira Silva, et al. (16). At first, DNA was extracted from each strain by boiling bacterial suspension of each isolate in TE buffer (Tris- EDTA), containing 2 Mercapthethanol (2%). Supernatant of boiling bacterial suspension after centrifugation was collected as a source of DNA. csgA and fimA genes specific primers were designed by Oliveira Silva (Table 1). Total volume of PCR reaction was 25 µL, which contained 12.5 µL of 2x PCR Master Mix (Ampliqon, Denmark), 1 µL (10 pmol/L) of each primer (forward and reverse), 5 µL of bacterial DNA, and 3.5 µL of nuclease-free water. PCR protocol was as follows: 1 cycle at 94°C for 4 minutes; 30 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 sseconds, followed by final extension step at 72°C for 4 minutes in a thermal cycler (Eppendorf, Germany). Escherichia coli (ATCC 25922) DNA and nuclease-free water were used as the positive and negative control, respectively. PCR products were electrophoresed and visualized in 1% agarose gel (Max Pure, Spain), and stained with safe stain (1X) (Cinaclon) using the UV transilluminator (UVtech- Germany).
| Gene | Primer | Length |
|---|---|---|
| csgA | 5’-ATCTGACCCAACGTGGCTTCG-3’ | 178 bp |
| 5’-GATGAGCGGTCGCGTTGTTACC-3’ | ||
| fimA | 5’-CTCTGGCAATCGTTGTTCTGTCG-3’ | 119 bp |
| 5’-GCAAGCGGCGTTAACAACTTCC-3’ |
Forward and Reverse Primers Used in csgA and fimA Genes Detection (16)
4. Results
In this study, after collecting 35 E. coli isolates by cultivation of 140 human urine samples, we evaluated the biofilm production ability of isolates by modified microtiter plates techniques described by Stepanovic et al. (15). Based on the results, 32 isolates (91.4%) were weak biofilm producers in BHI, containing 3% sucrose and only 1 isolate (2.8%) was moderate biofilm producer. In addition, 2 non- producing biofilm isolates (5.7%) in BHI medium were detected. None of the isolates was able to produce a strong level of biofilm in BHI, containing 3% sucrose medium. By PCR, 33 isolates (94.3%) were carriers for csgA gene, and 30 isolates (85.7%) were fimA gene carriers. Moreover, 5 and 2 samples were carriers for csgA and fimA genes, respectively (Figure 1). In this research, a correlation between presence of csgA and fimA genes and biofilm production ability was suspected because most of the isolates with and without biofilm production ability had csgA and fimA genes.
5. Discussion
Biofilm producer bacteria can cause a wide range of infections in humans and animals. Antibiotic and disinfectant agents’ resistance is 500 to 5000 times higher in biofilm producing bacteria than their planktonic form (17). Bacteria could be protected by expression of specific resistance genes in addition to production of large amount of exopolysaccharides under slow growing condition of biofilm production (18). The planktonic form of bacteria should be dispersed for colonization and biofilm production in new locations (19). The genotype of exointestinal E. coli strains could reflect the attachment ability of these strains to eukaryotic cells.
Several virulence factors presentations could help the organisms to adhere, colonize, or invade host cells, and facilitate a biofilm formation and create chronic illness. Adhesive organelles are commonly elaborated by UPEC, F1C, S, P, and Type I pili, encoded by the foc, sfa, pap, and fim operons, respectively (20). Type 1 and P pili, encoded by many UPEC strains, are the most studied adhesive organelles. P pili are often expressed in pyelonephritic UPEC isolates (21). The Fim H, which could mediate both invasion and bacterial adherence to host cells, contributes to intracellular bacterial biofilms formation by UPEC (22).
A variety of superficial and extracellular appendices such as flagella, Type I pili, and curli fimbriae are involved in biofilm formation of E. coli strains (23, 24). These superficial molecules could be detected by phenotypic and genotypic investigation. In fact, the biofilm synthesis regulation is highly complex, but little information is available on different species. Biofilm producing ability and correlation of different attachment factors were evaluated in several bacterial strains such as pathogenic Staphylococcus aureus (25) and E. coli (26-28).
Among different quantification methods for examining biofilm production ability in bacteria, polystyrene microtiter plate crystal violet system has been widely used (15, 29, 30) and biofilm production ability of different bacterial species has been investigated by this quantification method (14, 29, 31). In the present study, biofilm formation of 35 human urinary pathogen E. coli isolates were evaluated by microtiter plate crystal violet in BHI, containing 3% sucrose, and the correlation of presence of Type I fimbria and curli fimbria genes and biofilm ability in isolates were investigated using BHI containing 3% sucrose. In microtiter plate system, 94.3% of isolates were detected as biofilm producer, which is a significant percentage. In this medium, most of isolates (91.4%) were able to produce weak biofilm, and 2 isolates (5.7%) were not able to produce biofilm. In several studies, the effects of enrichment medium type in biofilm assay have been investigated. Samet et al. (32) and Stepanovic et al. (15) in separate studies introduced BHI medium better than others.
Samet et al. (32) in their study, used BHI containing 1% sucrose in biofilm production. In addition, because of the studied genes presence in most of isolates, with different ability in biofilm production (from moderate to no producing), no association was observed between biofilm producing ability and presence of csgA and fimA genes in uropathogenic E. coli strains. In Rijavec et al. (33) study, no association was observed between usp, papC, and sfa/foc virulence genes and biofilm production in pathogenic E. coli. However, higher frequency of papC, papG, sfa/foc, focG, hlyA and cnf1 genes in biofilm strong producer strains of E. coli were demonstrated by Naves et al. (34). Finally, due to the biofilm production by variable environmental factors, as same as type and species of bacteria, the diversity of UPEC-associated attachment factors with high levels of genetic similarity among nonpathogenic and pathogenic extraintestinal E. coli isolates makes it difficult to attribute biofilm producing ability to specially attachment factors.